Rational Design of Potent Peptide Inhibitors of the PD-1:PD-L1 Interaction for Cancer Immunotherapy.
Yin, H., Zhou, X., Huang, Y.H., King, G.J., Collins, B.M., Gao, Y., Craik, D.J., Wang, C.K.(2021) J Am Chem Soc 143: 18536-18547
- PubMed: 34661406
- DOI: https://doi.org/10.1021/jacs.1c08132
- Primary Citation of Related Structures:
7RJF - PubMed Abstract:
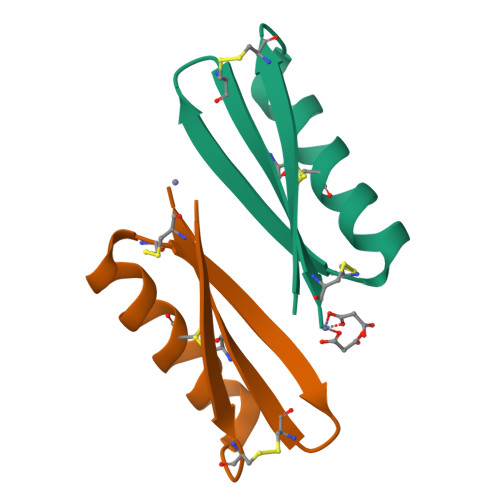
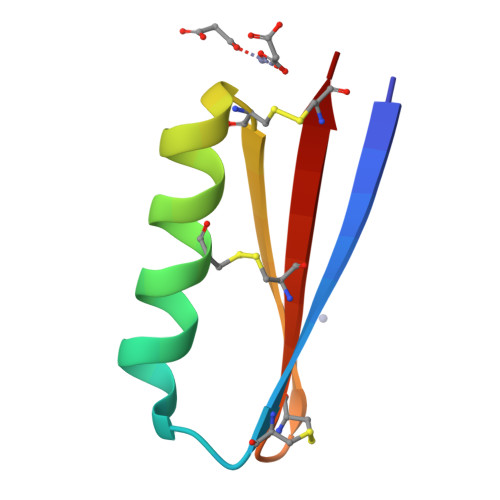
Peptides have potential to be developed into immune checkpoint inhibitors, but the target interfaces are difficult to inhibit. Here, we explored an approach to mimic the binding surface of PD-1 to design inhibitors. Mimicking native PD-1 resulted in a mimetic with no activity. However, mimicking an affinity-optimized PD-1 resulted in the peptide mimetic MOPD-1 that displayed nanomolar affinity to PD-L1 and could inhibit PD-1:PD-L1 interactions in both protein- and cell-based assays. Mutagenesis and structural characterization using NMR spectroscopy and X-ray crystallography revealed that binding residues from the high affinity PD-1 are crucial for the bioactivity of MOPD-1. Furthermore, MOPD-1 was extremely stable in human serum and inhibited tumor growth in vivo , suggesting it has potential for use in cancer immunotherapy. The successful design of an inhibitor of PD-1:PD-L1 using the mimicry approach described herein illustrates the value of placing greater emphasis on optimizing the target interface before inhibitor design and is an approach that could have broader utility for the design of peptide inhibitors for other complex protein-protein interactions.
Organizational Affiliation:
Institute for Molecular Bioscience, The University of Queensland, Brisbane, Queensland 4072, Australia.