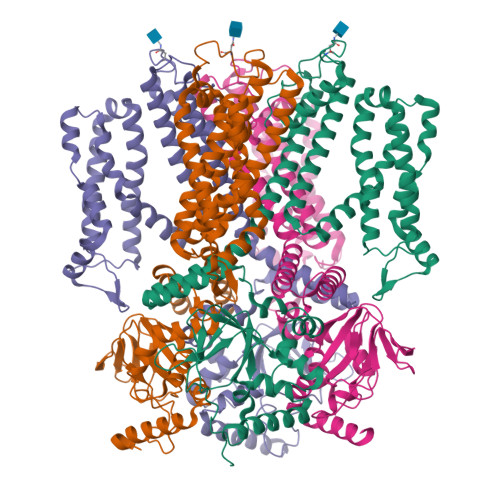
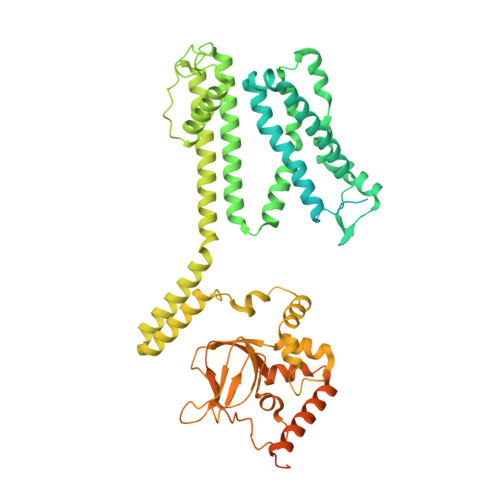
Structure of the human cone photoreceptor cyclic nucleotide-gated channel.
Zheng, X., Hu, Z., Li, H., Yang, J.(2022) Nat Struct Mol Biol 29: 40-46
- PubMed: 34969976
- DOI: https://doi.org/10.1038/s41594-021-00699-y
- Primary Citation of Related Structures:
7RHS - PubMed Abstract:
Cyclic nucleotide-gated (CNG) channels transduce light-induced chemical signals into electrical signals in retinal cone and rod photoreceptors. Structures of native CNG channels, which are heterotetramers formed by CNGA and CNGB subunits, have not been obtained. In the present study, we report a high-resolution cryo-electron microscopy structure of the human cone CNG channel in the apo closed state. The channel contains three CNGA3 and one CNGB3 subunits. Arg403 in the pore helix of CNGB3 projects into an asymmetric selectivity filter and forms hydrogen bonds with two pore-lining backbone carbonyl oxygens. Arg442 in S6 of CNGB3 protrudes into and occludes the pore below the hydrophobic cavity gate previously observed in homotetrameric CNGA channels. It is interesting that Arg403Gln is a disease mutation, and Arg442 is replaced by glutamine in some animal species with dichromatic or monochromatic vision. These and other unique structural features and the disease link conferred by CNGB3 indicate a critical role of CNGB3 in shaping cone photoresponses.
Organizational Affiliation:
Department of Biological Sciences, Columbia University, New York, NY, USA.