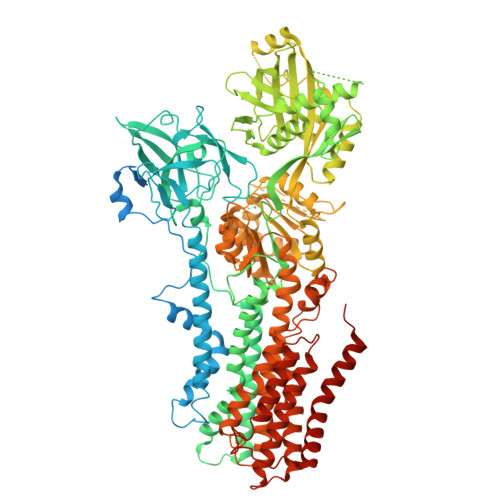
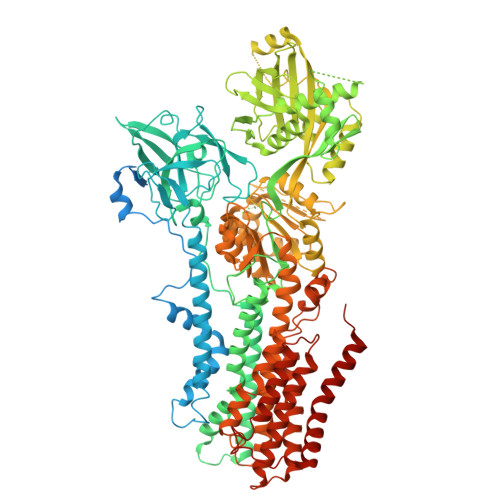
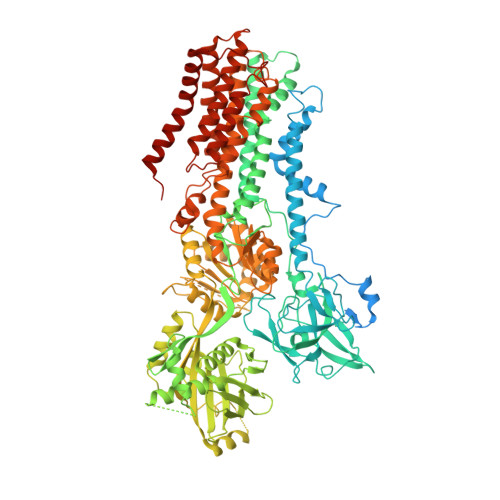
Structural basis of the P4B ATPase lipid flippase activity.
Bai, L., Jain, B.K., You, Q., Duan, H.D., Takar, M., Graham, T.R., Li, H.(2021) Nat Commun 12: 5963-5963
- PubMed: 34645814
- DOI: https://doi.org/10.1038/s41467-021-26273-0
- Primary Citation of Related Structures:
7RD6, 7RD7, 7RD8 - PubMed Abstract:
P4 ATPases are lipid flippases that are phylogenetically grouped into P4A, P4B and P4C clades. The P4A ATPases are heterodimers composed of a catalytic α-subunit and accessory β-subunit, and the structures of several heterodimeric flippases have been reported. The S. cerevisiae Neo1 and its orthologs represent the P4B ATPases, which function as monomeric flippases without a β-subunit. It has been unclear whether monomeric flippases retain the architecture and transport mechanism of the dimeric flippases. Here we report the structure of a P4B ATPase, Neo1, in its E1-ATP, E2P-transition, and E2P states. The structure reveals a conserved architecture as well as highly similar functional intermediate states relative to dimeric flippases. Consistently, structure-guided mutagenesis of residues in the proposed substrate translocation path disrupted Neo1's ability to establish membrane asymmetry. These observations indicate that evolutionarily distant P4 ATPases use a structurally conserved mechanism for substrate transport.
Organizational Affiliation:
Department of Biochemistry and Biophysics, School of Basic Medical Sciences, Peking University, Beijing, China. lbai@bjmu.edu.cn.