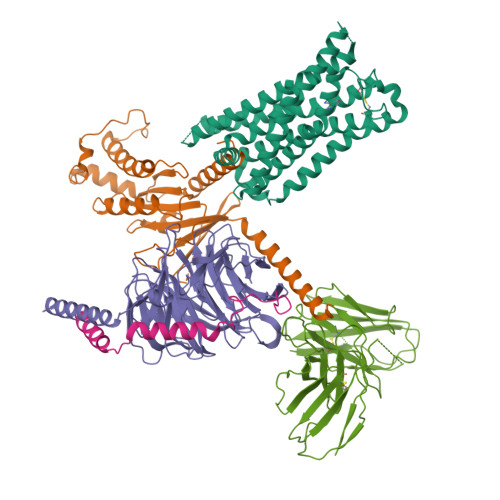
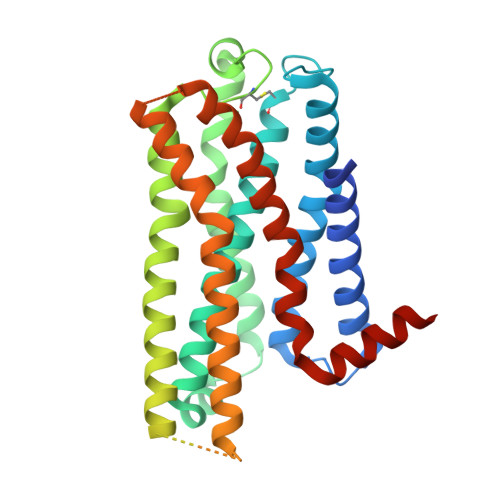
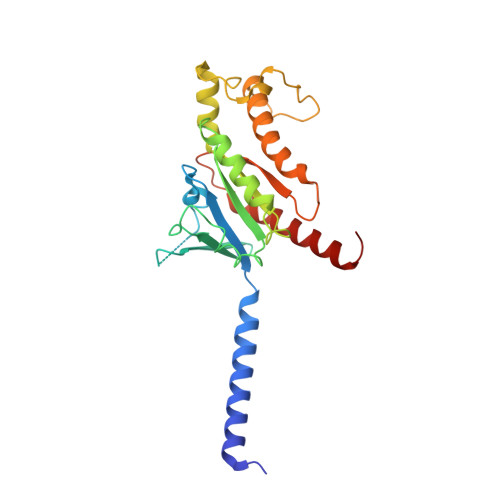
Bespoke library docking for 5-HT 2A receptor agonists with antidepressant activity.
Kaplan, A.L., Confair, D.N., Kim, K., Barros-Alvarez, X., Rodriguiz, R.M., Yang, Y., Kweon, O.S., Che, T., McCorvy, J.D., Kamber, D.N., Phelan, J.P., Martins, L.C., Pogorelov, V.M., DiBerto, J.F., Slocum, S.T., Huang, X.P., Kumar, J.M., Robertson, M.J., Panova, O., Seven, A.B., Wetsel, A.Q., Wetsel, W.C., Irwin, J.J., Skiniotis, G., Shoichet, B.K., Roth, B.L., Ellman, J.A.(2022) Nature 610: 582-591
- PubMed: 36171289
- DOI: https://doi.org/10.1038/s41586-022-05258-z
- Primary Citation of Related Structures:
7RAN - PubMed Abstract:
There is considerable interest in screening ultralarge chemical libraries for ligand discovery, both empirically and computationally 1-4 . Efforts have focused on readily synthesizable molecules, inevitably leaving many chemotypes unexplored. Here we investigate structure-based docking of a bespoke virtual library of tetrahydropyridines-a scaffold that is poorly sampled by a general billion-molecule virtual library but is well suited to many aminergic G-protein-coupled receptors. Using three inputs, each with diverse available derivatives, a one pot C-H alkenylation, electrocyclization and reduction provides the tetrahydropyridine core with up to six sites of derivatization 5-7 . Docking a virtual library of 75 million tetrahydropyridines against a model of the serotonin 5-HT 2A receptor (5-HT 2A R) led to the synthesis and testing of 17 initial molecules. Four of these molecules had low-micromolar activities against either the 5-HT 2A or the 5-HT 2B receptors. Structure-based optimization led to the 5-HT 2A R agonists (R)-69 and (R)-70, with half-maximal effective concentration values of 41 nM and 110 nM, respectively, and unusual signalling kinetics that differ from psychedelic 5-HT 2A R agonists. Cryo-electron microscopy structural analysis confirmed the predicted binding mode to 5-HT 2A R. The favourable physical properties of these new agonists conferred high brain permeability, enabling mouse behavioural assays. Notably, neither had psychedelic activity, in contrast to classic 5-HT 2A R agonists, whereas both had potent antidepressant activity in mouse models and had the same efficacy as antidepressants such as fluoxetine at as low as 1/40th of the dose. Prospects for using bespoke virtual libraries to sample pharmacologically relevant chemical space will be considered.
Organizational Affiliation:
Department of Pharmaceutical Chemistry, University of California, San Francisco, CA, USA.