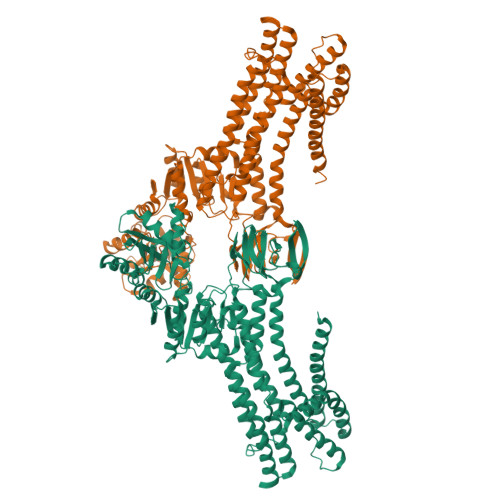
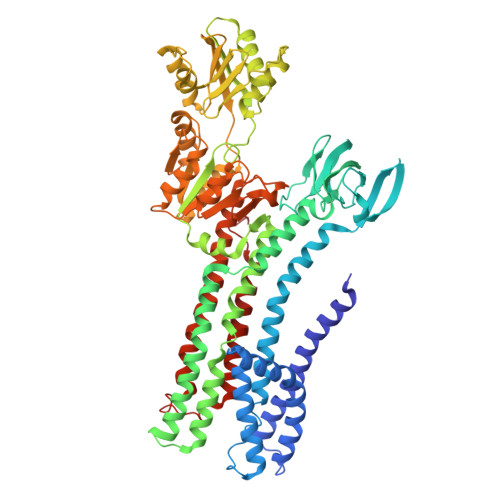
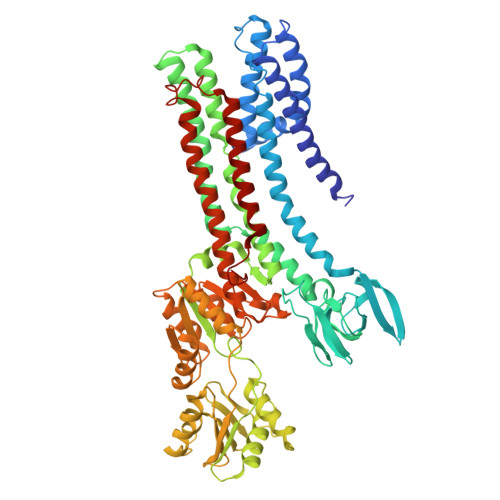
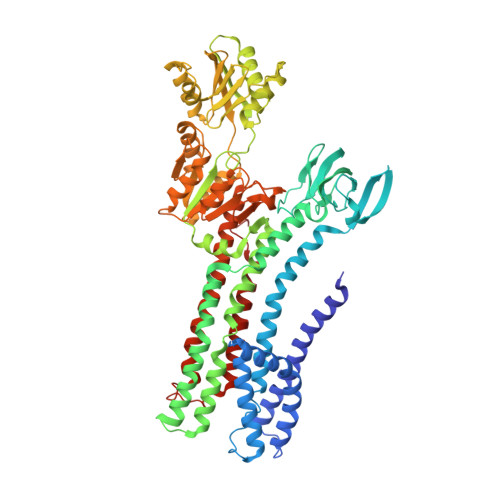
Structural basis of ion uptake in copper-transporting P 1B -type ATPases.
Salustros, N., Gronberg, C., Abeyrathna, N.S., Lyu, P., Oradd, F., Wang, K., Andersson, M., Meloni, G., Gourdon, P.(2022) Nat Commun 13: 5121-5121
- PubMed: 36045128
- DOI: https://doi.org/10.1038/s41467-022-32751-w
- Primary Citation of Related Structures:
7R0G, 7R0H, 7R0I - PubMed Abstract:
Copper is essential for living cells, yet toxic at elevated concentrations. Class 1B P-type (P 1B -) ATPases are present in all kingdoms of life, facilitating cellular export of transition metals including copper. P-type ATPases follow an alternating access mechanism, with inward-facing E1 and outward-facing E2 conformations. Nevertheless, no structural information on E1 states is available for P 1B -ATPases, hampering mechanistic understanding. Here, we present structures that reach 2.7 Å resolution of a copper-specific P 1B -ATPase in an E1 conformation, with complementing data and analyses. Our efforts reveal a domain arrangement that generates space for interaction with ion donating chaperones, and suggest a direct Cu + transfer to the transmembrane core. A methionine serves a key role by assisting the release of the chaperone-bound ion and forming a cargo entry site together with the cysteines of the CPC signature motif. Collectively, the findings provide insights into P 1B -mediated transport, likely applicable also to human P 1B -members.
Organizational Affiliation:
Department of Biomedical Sciences, Copenhagen University, Maersk Tower 7-9, Nørre Allé 14, DK-2200, Copenhagen, Denmark.