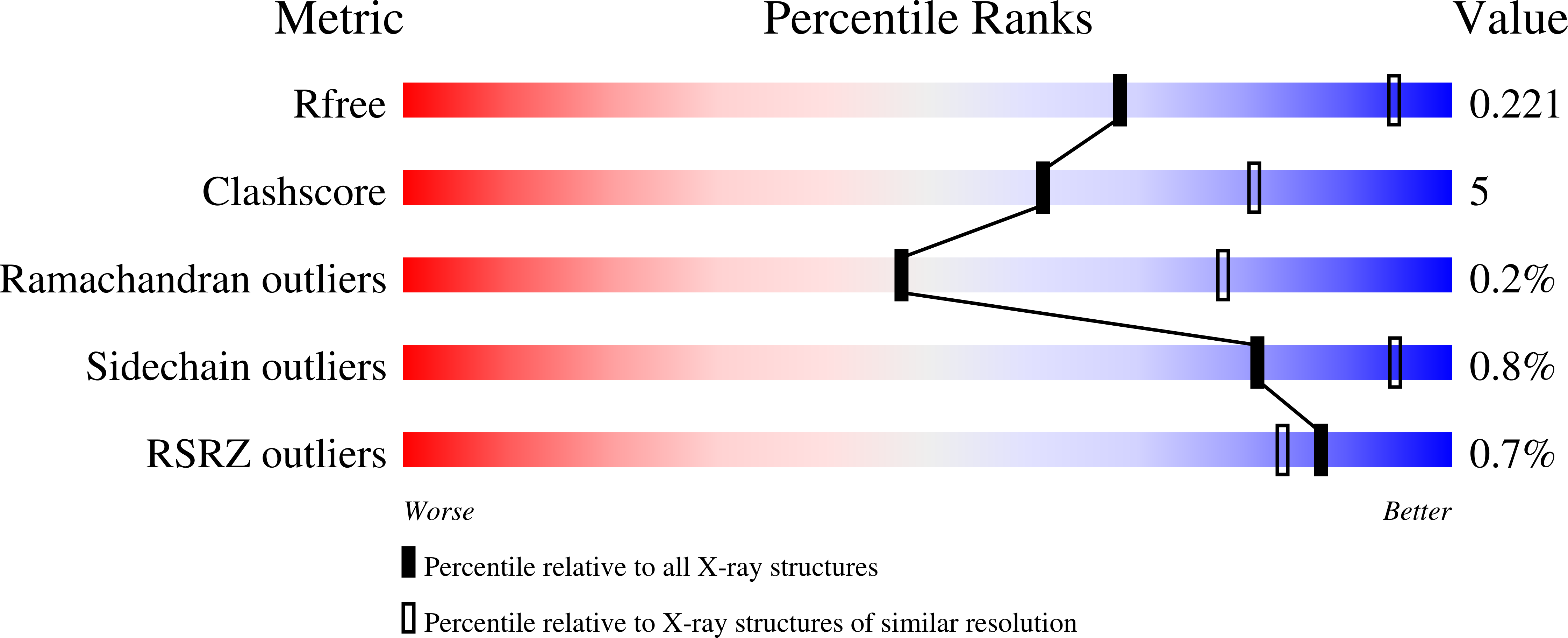
Broad-Spectrum Antidote Discovery by Untangling the Reactivation Mechanism of Nerve-Agent-Inhibited Acetylcholinesterase.
Lindgren, C., Forsgren, N., Hoster, N., Akfur, C., Artursson, E., Edvinsson, L., Svensson, R., Worek, F., Ekstrom, F., Linusson, A.(2022) Chemistry 28: e202200678-e202200678
- PubMed: 35420233
- DOI: https://doi.org/10.1002/chem.202200678
- Primary Citation of Related Structures:
7QYN, 7R02, 7R0A, 7R2F, 7R3C, 7R4E - PubMed Abstract:
Reactivators are vital for the treatment of organophosphorus nerve agent (OPNA) intoxication but new alternatives are needed due to their limited clinical applicability. The toxicity of OPNAs stems from covalent inhibition of the essential enzyme acetylcholinesterase (AChE), which reactivators relieve via a chemical reaction with the inactivated enzyme. Here, we present new strategies and tools for developing reactivators. We discover suitable inhibitor scaffolds by using an activity-independent competition assay to study non-covalent interactions with OPNA-AChEs and transform these inhibitors into broad-spectrum reactivators. Moreover, we identify determinants of reactivation efficiency by analysing reactivation and pre-reactivation kinetics together with structural data. Our results show that new OPNA reactivators can be discovered rationally by exploiting detailed knowledge of the reactivation mechanism of OPNA-inhibited AChE.
Organizational Affiliation:
Department of Chemistry, Umeå University, 901 87, Umeå, Sweden.