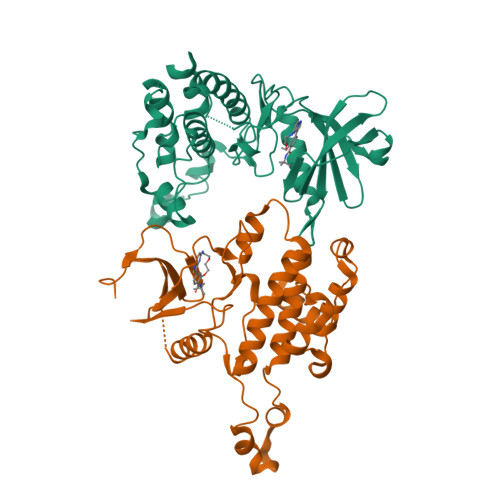
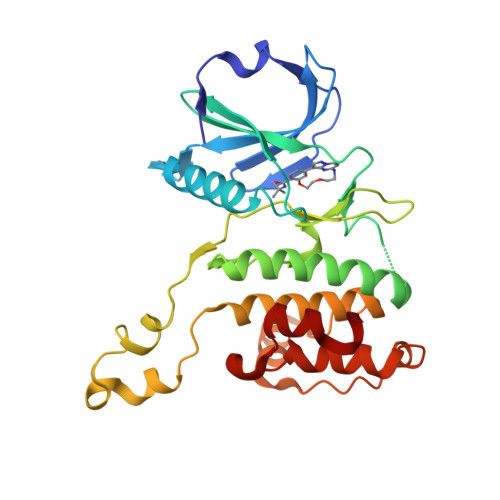
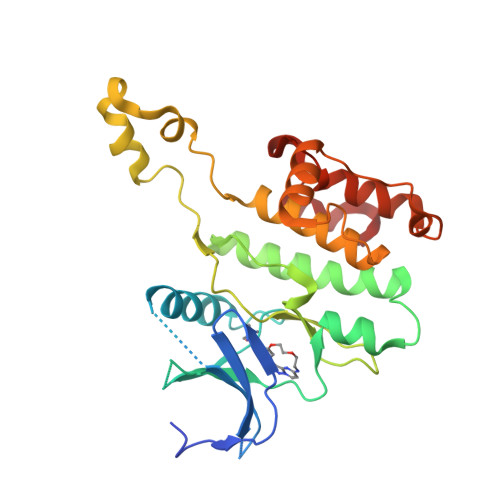
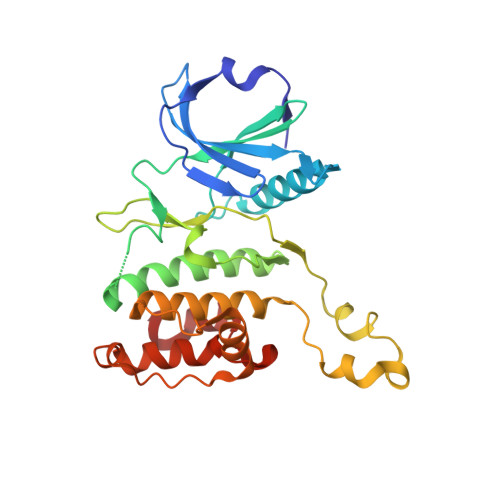
Illuminating the Dark: Highly Selective Inhibition of Serine/Threonine Kinase 17A with Pyrazolo[1,5- a ]pyrimidine-Based Macrocycles.
Kurz, C.G., Preuss, F., Tjaden, A., Cusack, M., Amrhein, J.A., Chatterjee, D., Mathea, S., Berger, L.M., Berger, B.T., Kramer, A., Weller, M., Weiss, T., Muller, S., Knapp, S., Hanke, T.(2022) J Med Chem 65: 7799-7817
- PubMed: 35608370
- DOI: https://doi.org/10.1021/acs.jmedchem.2c00173
- Primary Citation of Related Structures:
7QUE, 7QUF - PubMed Abstract:
Serine/threonine kinase 17A (death-associated protein kinase-related apoptosis-inducing protein kinase 1─DRAK1) is a part of the death-associated protein kinase (DAPK) family and belongs to the so-called dark kinome. Thus, the current state of knowledge of the cellular function of DRAK1 and its involvement in pathophysiological processes is very limited. Recently, DRAK1 has been implicated in tumorigenesis of glioblastoma multiforme (GBM) and other cancers, but no selective inhibitors of DRAK1 are available yet. To this end, we optimized a pyrazolo[1,5- a ]pyrimidine-based macrocyclic scaffold. Structure-guided optimization of this macrocyclic scaffold led to the development of CK156 ( 34 ), which displayed high in vitro potency ( K D = 21 nM) and selectivity in kinomewide screens. Crystal structures demonstrated that CK156 ( 34 ) acts as a type I inhibitor. However, contrary to studies using genetic knockdown of DRAK1, we have seen the inhibition of cell growth of glioma cells in 2D and 3D culture only at low micromolar concentrations.
Organizational Affiliation:
Institute of Pharmaceutical Chemistry, Goethe University Frankfurt, Max-von-Laue-Straße 9, Frankfurt 60438, Germany.