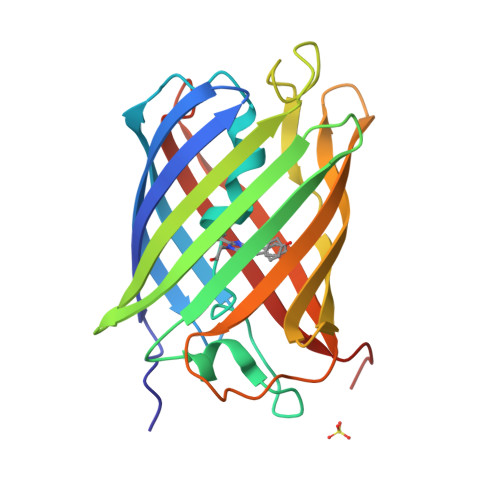
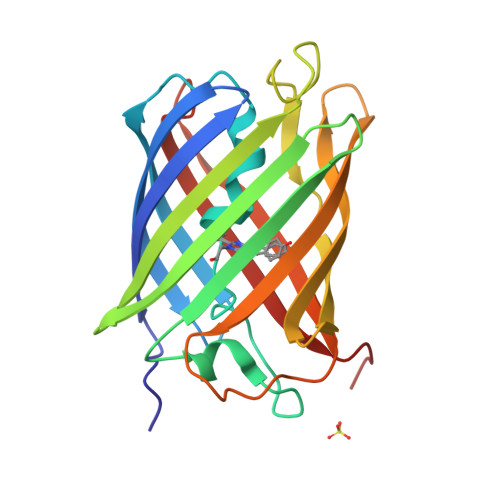
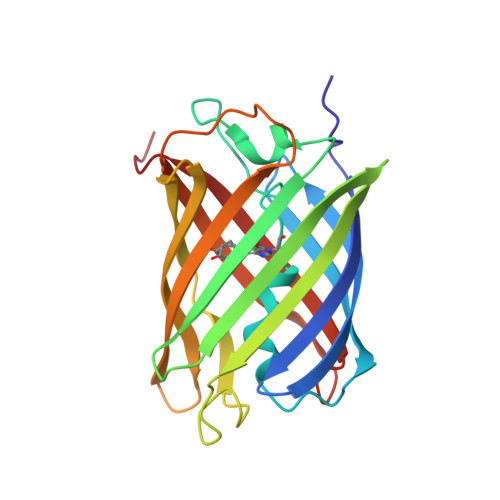
Optical control of ultrafast structural dynamics in a fluorescent protein.
Hutchison, C.D.M., Baxter, J.M., Fitzpatrick, A., Dorlhiac, G., Fadini, A., Perrett, S., Maghlaoui, K., Lefevre, S.B., Cordon-Preciado, V., Ferreira, J.L., Chukhutsina, V.U., Garratt, D., Barnard, J., Galinis, G., Glencross, F., Morgan, R.M., Stockton, S., Taylor, B., Yuan, L., Romei, M.G., Lin, C.Y., Marangos, J.P., Schmidt, M., Chatrchyan, V., Buckup, T., Morozov, D., Park, J., Park, S., Eom, I., Kim, M., Jang, D., Choi, H., Hyun, H., Park, G., Nango, E., Tanaka, R., Owada, S., Tono, K., DePonte, D.P., Carbajo, S., Seaberg, M., Aquila, A., Boutet, S., Barty, A., Iwata, S., Boxer, S.G., Groenhof, G., van Thor, J.J.(2023) Nat Chem 15: 1607-1615
- PubMed: 37563326
- DOI: https://doi.org/10.1038/s41557-023-01275-1
- Primary Citation of Related Structures:
7QLI, 7QLJ, 7QLK, 7QLL, 7QLM, 7QLN, 7QLO - PubMed Abstract:
The photoisomerization reaction of a fluorescent protein chromophore occurs on the ultrafast timescale. The structural dynamics that result from femtosecond optical excitation have contributions from vibrational and electronic processes and from reaction dynamics that involve the crossing through a conical intersection. The creation and progression of the ultrafast structural dynamics strongly depends on optical and molecular parameters. When using X-ray crystallography as a probe of ultrafast dynamics, the origin of the observed nuclear motions is not known. Now, high-resolution pump-probe X-ray crystallography reveals complex sub-ångström, ultrafast motions and hydrogen-bonding rearrangements in the active site of a fluorescent protein. However, we demonstrate that the measured motions are not part of the photoisomerization reaction but instead arise from impulsively driven coherent vibrational processes in the electronic ground state. A coherent-control experiment using a two-colour and two-pulse optical excitation strongly amplifies the X-ray crystallographic difference density, while it fully depletes the photoisomerization process. A coherent control mechanism was tested and confirmed the wave packets assignment.
Organizational Affiliation:
Department of Life Sciences, Faculty of Natural Sciences, Imperial College London, London, UK.