Identification and optimisation of a pyrimidopyridone series of IRAK4 inhibitors.
Cumming, I.A., Degorce, S.L., Aagaard, A., Braybrooke, E.L., Davies, N.L., Diene, C.R., Eatherton, A.J., Felstead, H.R., Groombridge, S.D., Lenz, E.M., Li, Y., Nai, Y., Pearson, S., Robb, G.R., Scott, J.S., Steward, O.R., Wu, C., Xue, Y., Zhang, L., Zhang, Y.(2022) Bioorg Med Chem 63: 116729-116729
- PubMed: 35439688
- DOI: https://doi.org/10.1016/j.bmc.2022.116729
- Primary Citation of Related Structures:
7QG1, 7QG2, 7QG3, 7QG5 - PubMed Abstract:
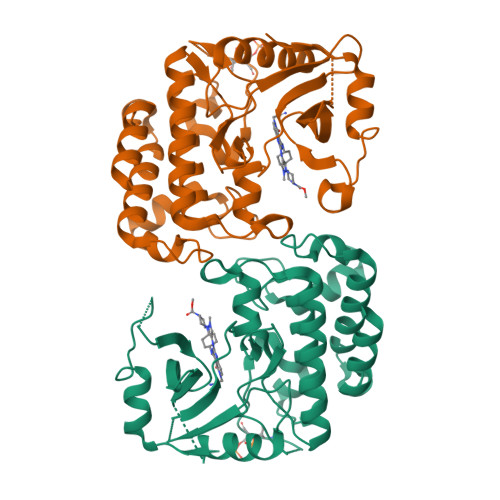
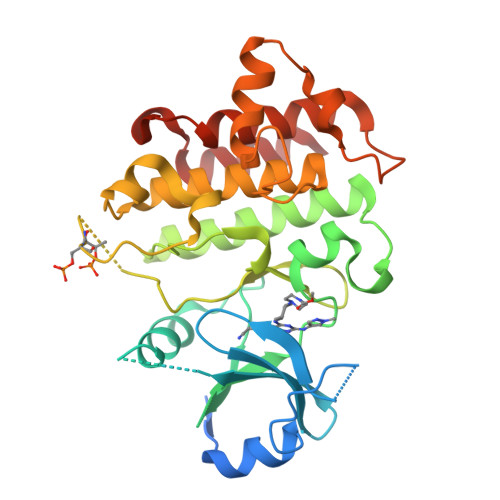
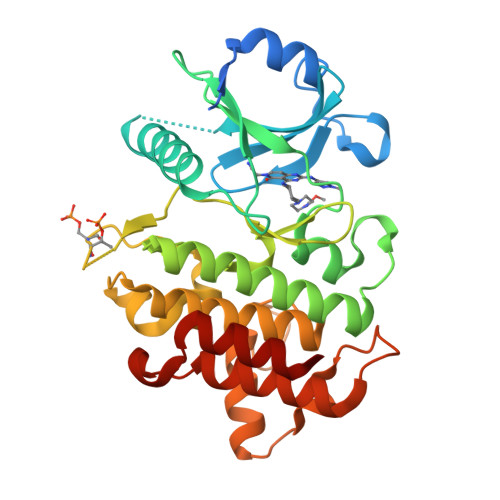
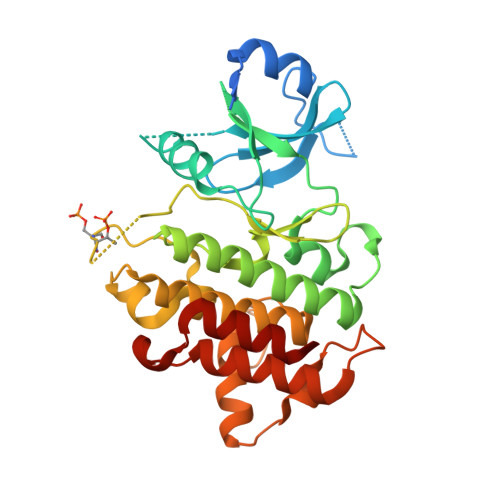
In this article, we report the discovery of a series of pyrimidopyridones as inhibitors of IRAK4 kinase. From a previously disclosed 5-azaquinazoline series, we found that switching the pyridine ring for an N-substituted pyridone gave a novel hinge binding scaffold which retained potency against IRAK4. Importantly, introduction of the carbonyl established an internal hydrogen bond with the 4-NH, establishing a conformational lock and allowing truncation of the large basic substituent to a 1-methylcyclopyl group. Subsequent optimisation, facilitated by X-ray crystal structures, allowed identification of preferred substituents at both the pyridone core and pyrazole. Subsequent combinations of optimal groups allowed control of lipophilicity and identification of potent and selective inhibitors of IRAK4 with better in vitro permeability and lower clearance.
Organizational Affiliation:
Medicinal Chemistry, Research and Early Development, Oncology R&D, AstraZeneca, Cambridge Science Park, Unit 310 Darwin Building, Cambridge CB4 0WG, United Kingdom. Electronic address: iain.cumming@astrazeneca.com.