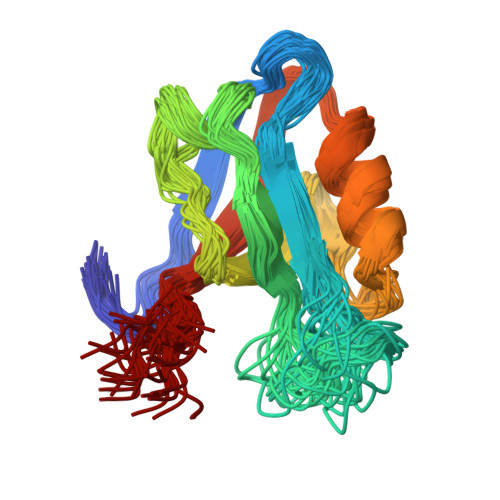
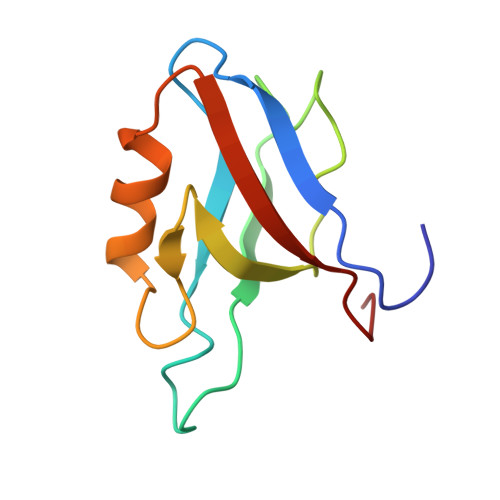
Atomic resolution protein allostery from the multi-state structure of a PDZ domain.
Ashkinadze, D., Kadavath, H., Pokharna, A., Chi, C.N., Friedmann, M., Strotz, D., Kumari, P., Minges, M., Cadalbert, R., Konigl, S., Guntert, P., Vogeli, B., Riek, R.(2022) Nat Commun 13: 6232-6232
- PubMed: 36266302
- DOI: https://doi.org/10.1038/s41467-022-33687-x
- Primary Citation of Related Structures:
7QCX, 7QCY - PubMed Abstract:
Recent methodological advances in solution NMR allow the determination of multi-state protein structures and provide insights into structurally and dynamically correlated protein sites at atomic resolution. This is demonstrated in the present work for the well-studied PDZ2 domain of protein human tyrosine phosphatase 1E for which protein allostery had been predicted. Two-state protein structures were calculated for both the free form and in complex with the RA-GEF2 peptide using the exact nuclear Overhauser effect (eNOE) method. In the apo protein, an allosteric conformational selection step comprising almost 60% of the domain was detected with an "open" ligand welcoming state and a "closed" state that obstructs the binding site by changing the distance between the β-sheet 2, α-helix 2, and sidechains of residues Lys38 and Lys72. The observed induced fit-type apo-holo structural rearrangements are in line with the previously published evolution-based analysis covering ~25% of the domain with only a partial overlap with the protein allostery of the open form. These presented structural studies highlight the presence of a dedicated highly optimized and complex dynamic interplay of the PDZ2 domain owed by the structure-dynamics landscape.
Organizational Affiliation:
Laboratory of Physical Chemistry, ETH Zürich, Vladimir-Prelog-Weg 2, CH-8093, Zürich, Switzerland.