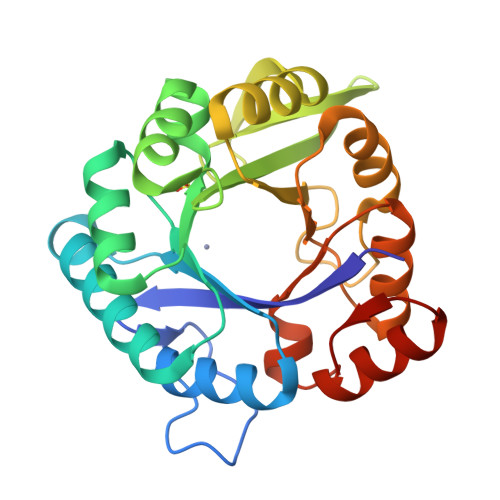
An artificial metalloprotein with metal-adaptive coordination sites and Ni-dependent quercetinase activity.
Beaumet, M., Dose, A., Brauer, A., Mahy, J.P., Ghattas, W., Groll, M., Hess, C.R.(2022) J Inorg Biochem 235: 111914-111914
- PubMed: 35841720
- DOI: https://doi.org/10.1016/j.jinorgbio.2022.111914
- Primary Citation of Related Structures:
7QC3, 7QC6, 7QC7, 7QC8, 7QC9 - PubMed Abstract:
Engineering non-native metal active sites into proteins using canonical amino acids offers many advantages but is hampered by significant challenges. The TIM barrel protein, imidazole glycerol phosphate synthase from the hyperthermophilic organism Thermotoga maritima (tHisF), is well-suited for the construction of artificial metalloenzymes by this approach. To this end, we have generated a tHisF variant (tHisF EHH ) with a Glu/His/His motif for metal ion coordination. Crystal structures of Zn II :tHisF EHH and Ni II :tHisF EHH reveal that both metal ions bind to the engineered histidines. However, the two metals bind at distinct sites with different geometries, demonstrating the adaptability of tHisF. Only Zn II additionally ligates the Glu residue and adopts a tetrahedral geometry. The pseudo-octahedral Ni II site comprises the two His and a native Ser residue. Ni II :tHisF EHH catalyzes the oxidative cleavage of the flavanols quercetin and myricetin, providing an unprecedented example of an artificial metalloprotein with quercetinase activity.
Organizational Affiliation:
Institut de Chimie Moléculaire et des Matériaux d'Orsay, Université Paris-Saclay, CNRS, 91405 Orsay, France.