Highly potent inhibitors of cathepsin K with a differently positioned cyanohydrazide warhead: structural analysis of binding mode to mature and zymogen-like enzymes.
Benysek, J., Busa, M., Rubesova, P., Fanfrlik, J., Lepsik, M., Brynda, J., Matouskova, Z., Bartz, U., Horn, M., Gutschow, M., Mares, M.(2022) J Enzyme Inhib Med Chem 37: 515-526
- PubMed: 35144520
- DOI: https://doi.org/10.1080/14756366.2021.2024527
- Primary Citation of Related Structures:
7QBL, 7QBM, 7QBN, 7QBO - PubMed Abstract:
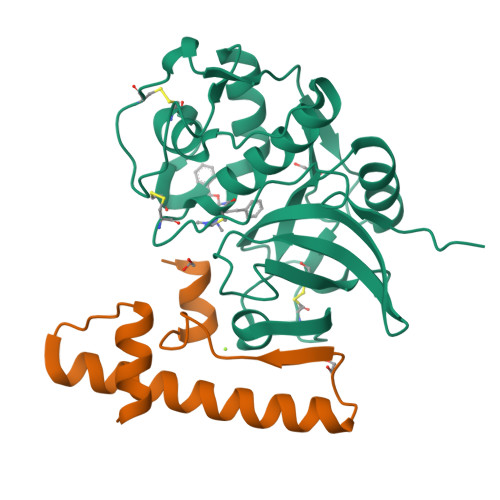
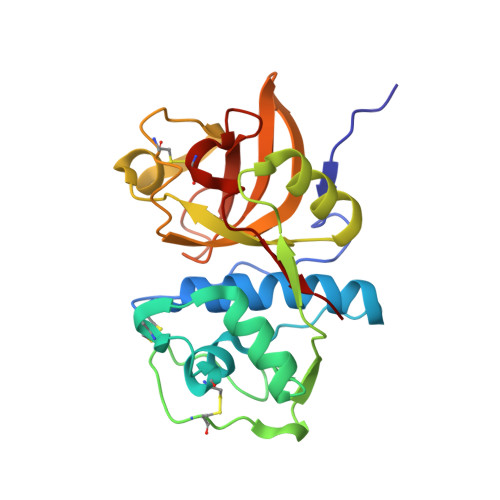
Cathepsin K (CatK) is a target for the treatment of osteoporosis, arthritis, and bone metastasis. Peptidomimetics with a cyanohydrazide warhead represent a new class of highly potent CatK inhibitors; however, their binding mechanism is unknown. We investigated two model cyanohydrazide inhibitors with differently positioned warheads: an azadipeptide nitrile Gü1303 and a 3-cyano-3-aza-β-amino acid Gü2602 . Crystal structures of their covalent complexes were determined with mature CatK as well as a zymogen-like activation intermediate of CatK. Binding mode analysis, together with quantum chemical calculations, revealed that the extraordinary picomolar potency of Gü2602 is entropically favoured by its conformational flexibility at the nonprimed-primed subsites boundary. Furthermore, we demonstrated by live cell imaging that cyanohydrazides effectively target mature CatK in osteosarcoma cells. Cyanohydrazides also suppressed the maturation of CatK by inhibiting the autoactivation of the CatK zymogen. Our results provide structural insights for the rational design of cyanohydrazide inhibitors of CatK as potential drugs.
Organizational Affiliation:
Institute of Organic Chemistry and Biochemistry of the Czech Academy of Sciences, Prague, Czech Republic.