Generation of nanobodies targeting the human, transcobalamin-mediated vitamin B 12 uptake route.
Bloch, J.S., Sequeira, J.M., Ramirez, A.S., Quadros, E.V., Locher, K.P.(2022) FASEB J 36: e22222-e22222
- PubMed: 35218573
- DOI: https://doi.org/10.1096/fj.202101376RR
- Primary Citation of Related Structures:
7QBD, 7QBE, 7QBF, 7QBG - PubMed Abstract:
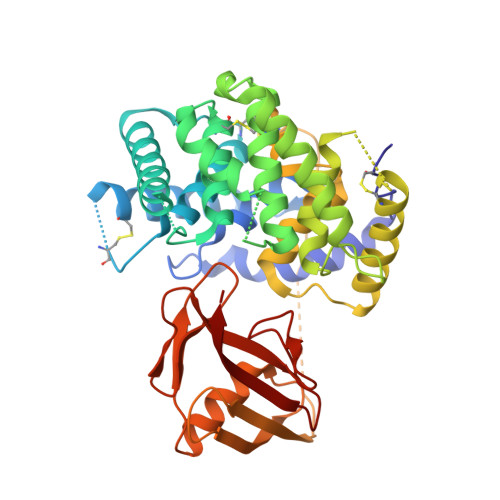
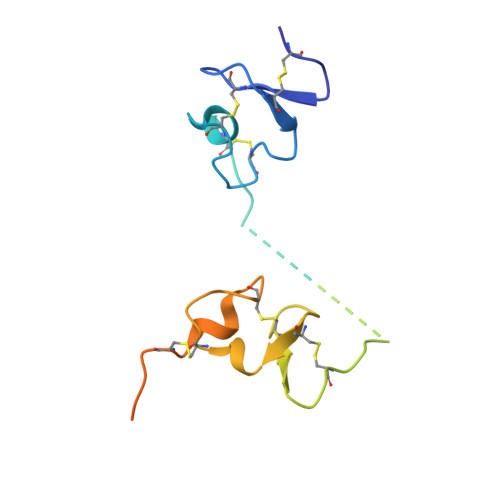
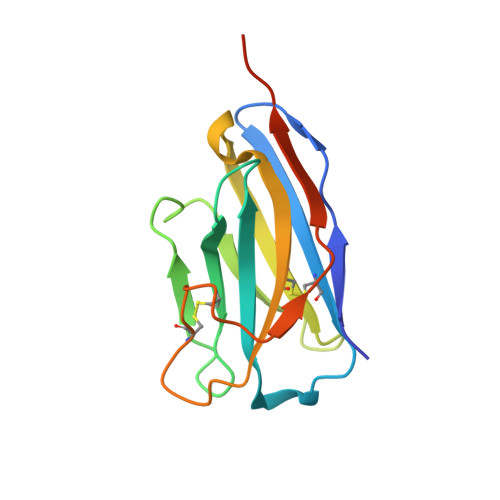
Cellular uptake of vitamin B 12 in humans is mediated by the endocytosis of the B 12 carrier protein transcobalamin (TC) via its cognate cell surface receptor TCblR, encoded by the CD320 gene. Because CD320 expression is associated with the cell cycle and upregulated in highly proliferating cells including cancer cells, this uptake route is a potential target for cancer therapy. We developed and characterized four camelid nanobodies that bind holo-TC (TC in complex with B 12 ) or the interface of the human holo-TC:TCblR complex with nanomolar affinities. We determined X-ray crystal structures of these nanobodies bound to holo-TC:TCblR, which enabled us to map their binding epitopes. When conjugated to the model toxin saporin, three of our nanobodies caused growth inhibition of HEK293T cells and therefore have the potential to inhibit the growth of human cancer cells. We visualized the cellular binding and endocytic uptake of the most potent nanobody (TC-Nb4) using fluorescent light microscopy. The co-crystal structure of holo-TC:TCblR with another nanobody (TC-Nb34) revealed novel features of the interface of TC and the LDLR-A1 domain of TCblR, rationalizing the decrease in the affinity of TC-B 12 binding caused by the Δ88 mutation in CD320.
Organizational Affiliation:
Institute of Molecular Biology and Biophysics, ETH Zürich, Zürich, Switzerland.