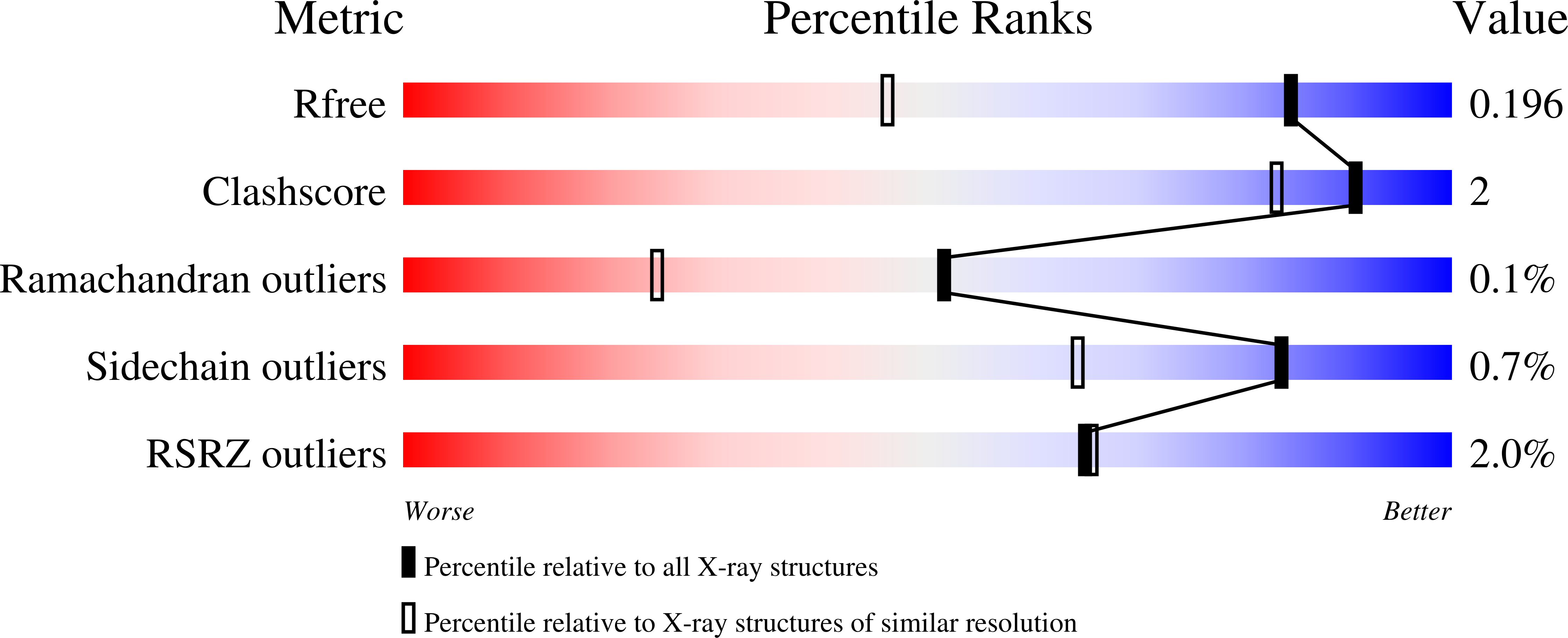
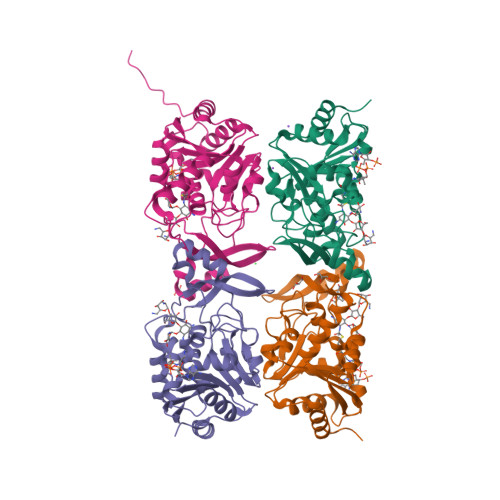
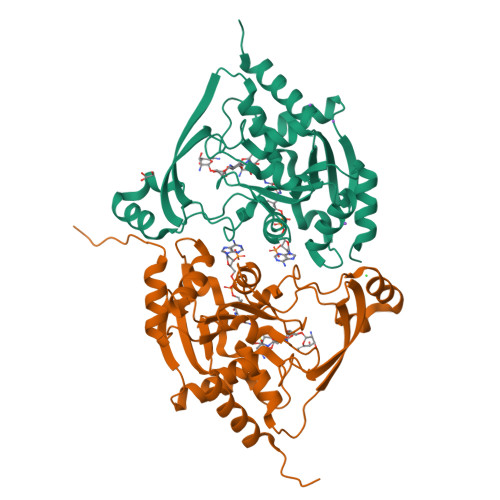
3-N-alkylation in aminoglycoside antibiotic neomycin B overcomes bacterial resistance mediated by acetyltransferase (3) IIIa
Pontillo, N., Warszawik, E.M., Partipilo, M., Stetsenko, A., Loznik, M., Yang, X., Slotboom, D.J., Guskov, A., Herrmann, A.To be published.