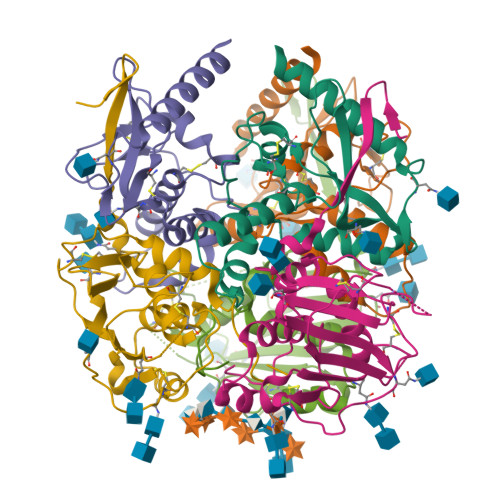
Structure and receptor recognition by the Lassa virus spike complex.
Katz, M., Weinstein, J., Eilon-Ashkenazy, M., Gehring, K., Cohen-Dvashi, H., Elad, N., Fleishman, S.J., Diskin, R.(2022) Nature 603: 174-179
- PubMed: 35173332
- DOI: https://doi.org/10.1038/s41586-022-04429-2
- Primary Citation of Related Structures:
7PUY, 7PVD - PubMed Abstract:
Lassa virus (LASV) is a human pathogen, causing substantial morbidity and mortality 1,2 . Similar to other Arenaviridae, it presents a class-I spike complex on its surface that facilitates cell entry. The virus's cellular receptor is matriglycan, a linear carbohydrate that is present on α-dystroglycan 3,4 , but the molecular mechanism that LASV uses to recognize this glycan is unknown. In addition, LASV and other arenaviruses have a unique signal peptide that forms an integral and functionally important part of the mature spike 5-8 ; yet the structure, function and topology of the signal peptide in the membrane remain uncertain 9-11 . Here we solve the structure of a complete native LASV spike complex, finding that the signal peptide crosses the membrane once and that its amino terminus is located in the extracellular region. Together with a double-sided domain-switching mechanism, the signal peptide helps to stabilize the spike complex in its native conformation. This structure reveals that the LASV spike complex is preloaded with matriglycan, suggesting the mechanism of binding and rationalizing receptor recognition by α-dystroglycan-tropic arenaviruses. This discovery further informs us about the mechanism of viral egress and may facilitate the rational design of novel therapeutics that exploit this binding site.
Organizational Affiliation:
Department of Chemical and Structural Biology, Weizmann Institute of Science, Rehovot, Israel.