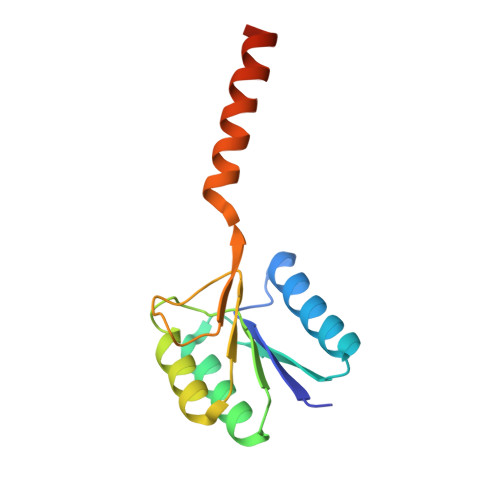
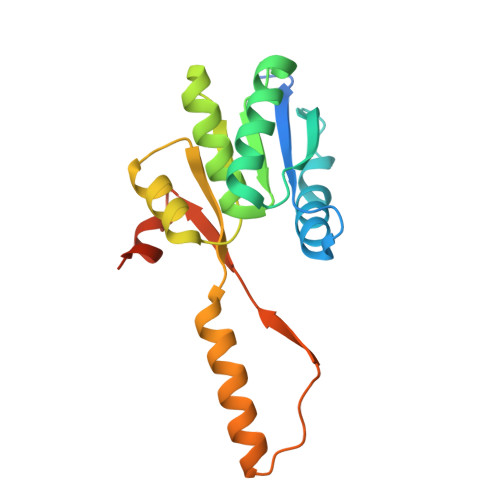
Retracing the evolution of a modern periplasmic binding protein.
Michel, F., Romero-Romero, S., Hocker, B.(2023) Protein Sci : e4793-e4793
- PubMed: 37788980
- DOI: https://doi.org/10.1002/pro.4793
- Primary Citation of Related Structures:
7PU4 - PubMed Abstract:
Investigating the evolution of structural features in modern multidomain proteins helps to understand their immense diversity and functional versatility. The class of periplasmic binding proteins (PBPs) offers an opportunity to interrogate one of the main processes driving diversification: the duplication and fusion of protein sequences to generate new architectures. The symmetry of their two-lobed topology, their mechanism of binding, and the organization of their operon structure led to the hypothesis that PBPs arose through a duplication and fusion event of a single common ancestor. To investigate this claim, we set out to reverse the evolutionary process and recreate the structural equivalent of a single-lobed progenitor using ribose-binding protein (RBP) as our model. We found that this modern PBP can be deconstructed into its lobes, producing two proteins that represent possible progenitor halves. The isolated halves of RBP are well folded and monomeric proteins, albeit with a lower thermostability, and do not retain the original binding function. However, the two entities readily form a heterodimer in vitro and in-cell. The x-ray structure of the heterodimer closely resembles the parental protein. Moreover, the binding function is fully regained upon formation of the heterodimer with a ligand affinity similar to that observed in the modern RBP. This highlights how a duplication event could have given rise to a stable and functional PBP-like fold and provides insights into how more complex functional structures can evolve from simpler molecular components.
Organizational Affiliation:
Department of Biochemistry, University of Bayreuth, Bayreuth, Germany.