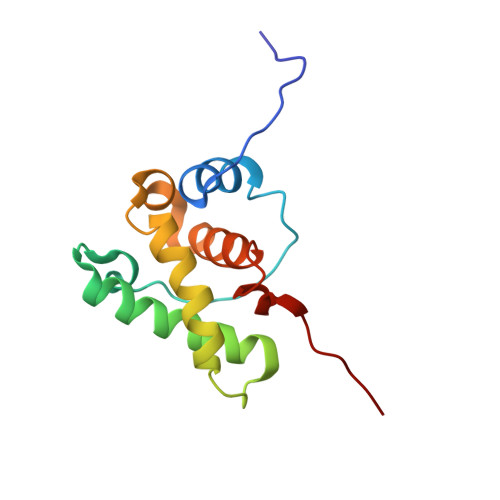
Structural basis of Nrd1-Nab3 heterodimerization.
Chaves-Arquero, B., Martinez-Lumbreras, S., Camero, S., Santiveri, C.M., Mirassou, Y., Campos-Olivas, R., Jimenez, M.A., Calvo, O., Perez-Canadillas, J.M.(2022) Life Sci Alliance 5
- PubMed: 35022249
- DOI: https://doi.org/10.26508/lsa.202101252
- Primary Citation of Related Structures:
7PRD, 7PRE - PubMed Abstract:
Heterodimerization of RNA binding proteins Nrd1 and Nab3 is essential to communicate the RNA recognition in the nascent transcript with the Nrd1 recognition of the Ser 5 -phosphorylated Rbp1 C-terminal domain in RNA polymerase II. The structure of a Nrd1-Nab3 chimera reveals the basis of heterodimerization, filling a missing gap in knowledge of this system. The free form of the Nrd1 interaction domain of Nab3 (NRID) forms a multi-state three-helix bundle that is clamped in a single conformation upon complex formation with the Nab3 interaction domain of Nrd1 (NAID). The latter domain forms two long helices that wrap around NRID, resulting in an extensive protein-protein interface that would explain the highly favorable free energy of heterodimerization. Mutagenesis of some conserved hydrophobic residues involved in the heterodimerization leads to temperature-sensitive phenotypes, revealing the importance of this interaction in yeast cell fitness. The Nrd1-Nab3 structure resembles the previously reported Rna14/Rna15 heterodimer structure, which is part of the poly(A)-dependent termination pathway, suggesting that both machineries use similar structural solutions despite they share little sequence homology and are potentially evolutionary divergent.
Organizational Affiliation:
Departamento de Química-Física Biológica, Instituto de Química-Física "Rocasolano" (IQFR), Consejo Superior de Investigaciones Científicas (CSIC), Madrid, Spain.