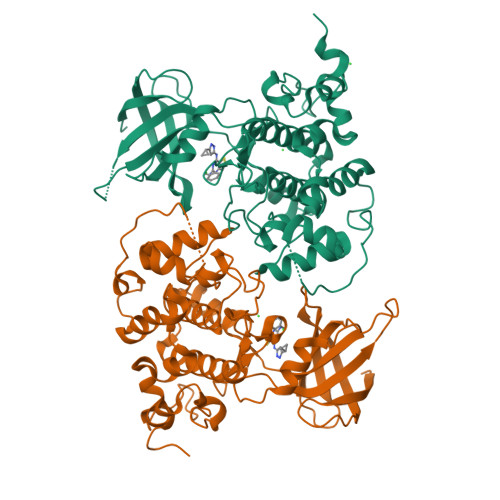
ARN25068, a versatile starting point towards triple GSK-3 beta /FYN/DYRK1A inhibitors to tackle tau-related neurological disorders.
Demuro, S., Sauvey, C., Tripathi, S.K., Di Martino, R.M.C., Shi, D., Ortega, J.A., Russo, D., Balboni, B., Giabbai, B., Storici, P., Girotto, S., Abagyan, R., Cavalli, A.(2022) Eur J Med Chem 229: 114054-114054
- PubMed: 34959172
- DOI: https://doi.org/10.1016/j.ejmech.2021.114054
- Primary Citation of Related Structures:
7OY5, 7OY6 - PubMed Abstract:
The human kinome plays a crucial role in several pathways. Its dysregulation has been linked to diverse central nervous system (CNS)-related disorders with a drastic impact on the aging population. Among them, tauopathies, such as Alzheimer's Disease (AD) and Frontotemporal Lobar degeneration (FTLD-tau), are neurodegenerative disorders pathologically defined by the presence of hyperphosphorylated tau-positive intracellular inclusions known as neurofibrillary tangles (NFTs). Compelling evidence has reported the great potential of the simultaneous modulation of multiple protein kinases (PKs) involved in abnormal tau phosphorylation through a concerted pharmacological approach to achieve a superior therapeutic effect relative to classic "one target, one drug" approaches. Here, we report on the identification and characterization of ARN25068 (4), a low nanomolar and well-balanced dual GSK-3β and FYN inhibitor, which also shows inhibitory activity against DYRK1A, an emerging target in AD and tauopathies. Computational and X-Ray studies highlight compound 4's typical H-bonding pattern of ATP-competitive inhibitors at the binding sites of all three PKs. In a tau phosphorylation assay on Tau0N4R-TM-tGFP U2OS cell line, 4 reduces the extent of tau phosphorylation, promoting tau-stabilized microtubule bundles. In conclusion, this compound emerges as a promising prototype for further SAR explorations to develop potent and well-balanced triple GSK-3β/FYN/DYRK1A inhibitors to tackle tau hyperphosphorylation.
Organizational Affiliation:
Computational and Chemical Biology, Istituto Italiano di Tecnologia, Via Morego 30, 16163, Genoa, Italy; Department of Pharmacy and Biotechnology, University of Bologna, Via Belmeloro 6, 40126, Bologna, Italy.