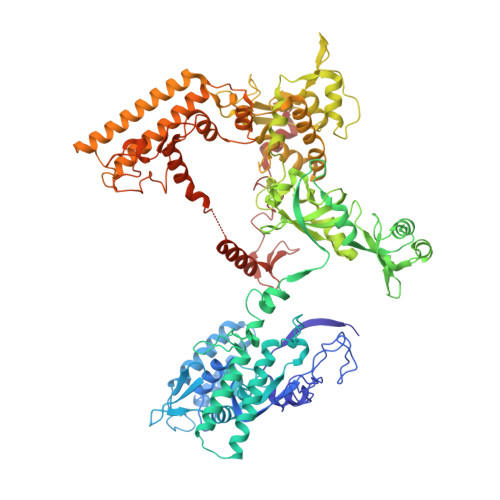
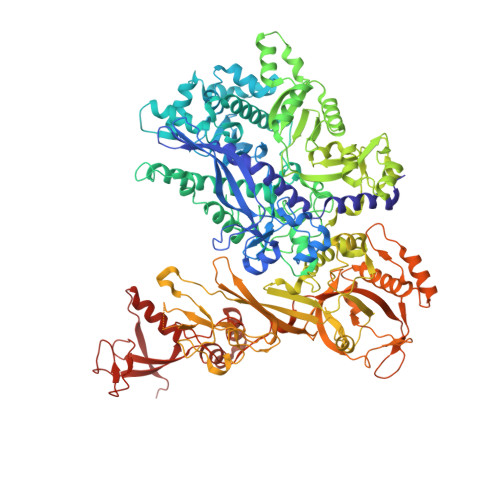
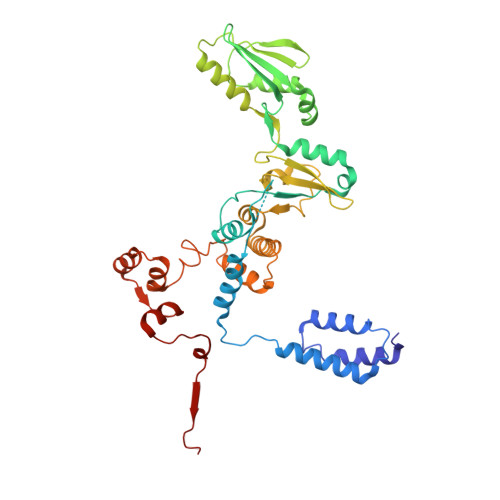
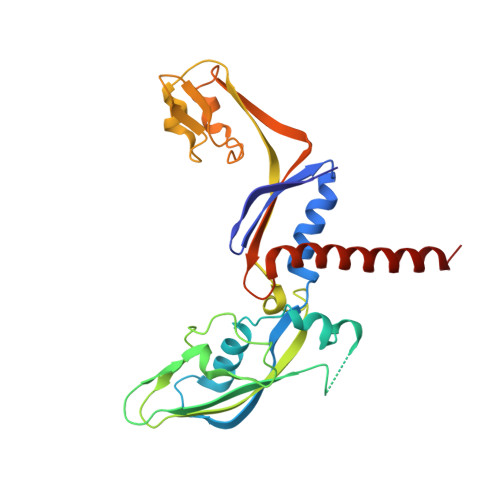
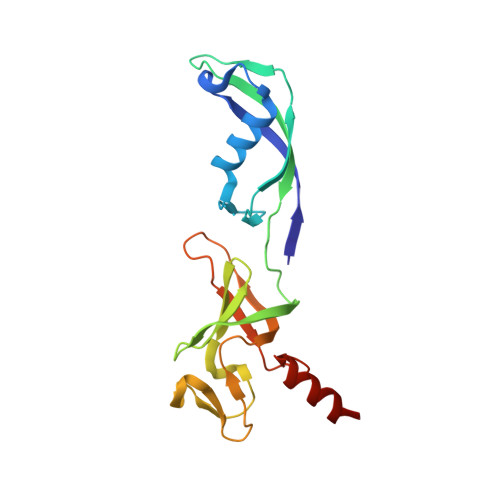
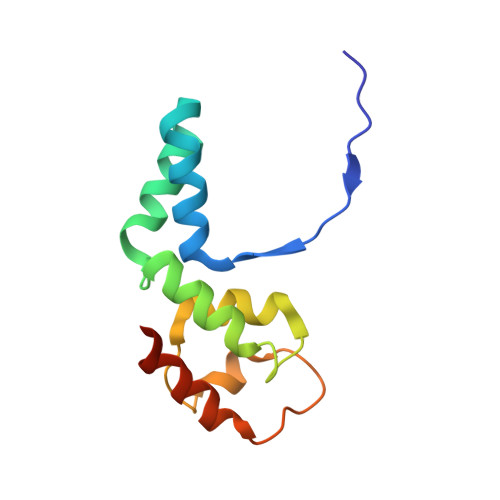
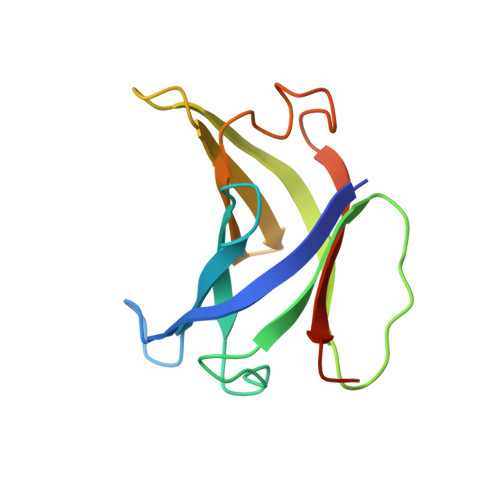
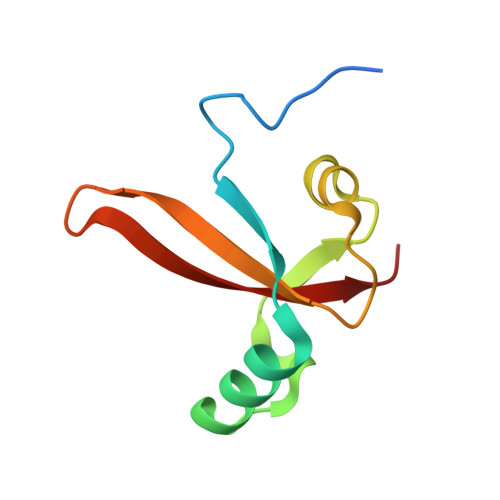
Structural basis of RNA polymerase inhibition by viral and host factors.
Pilotto, S., Fouqueau, T., Lukoyanova, N., Sheppard, C., Lucas-Staat, S., Diaz-Santin, L.M., Matelska, D., Prangishvili, D., Cheung, A.C.M., Werner, F.(2021) Nat Commun 12: 5523-5523
- PubMed: 34535646
- DOI: https://doi.org/10.1038/s41467-021-25666-5
- Primary Citation of Related Structures:
7OK0, 7OQ4, 7OQY - PubMed Abstract:
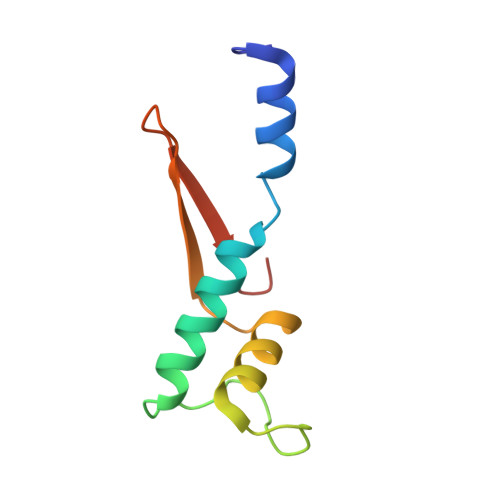
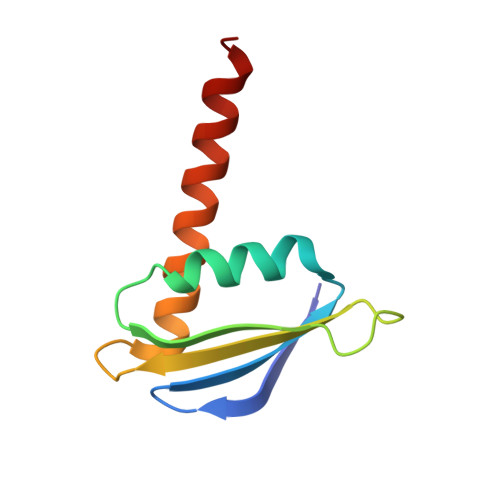
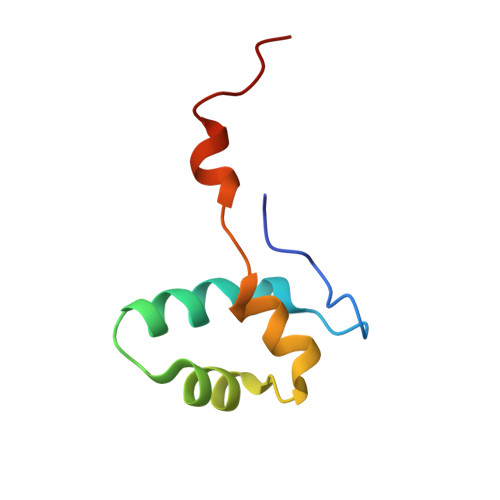
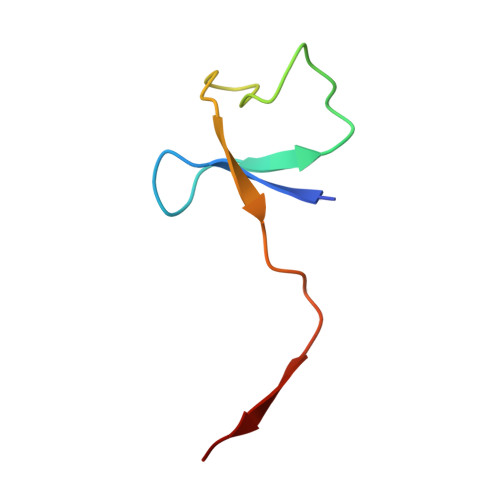
RNA polymerase inhibition plays an important role in the regulation of transcription in response to environmental changes and in the virus-host relationship. Here we present the high-resolution structures of two such RNAP-inhibitor complexes that provide the structural bases underlying RNAP inhibition in archaea. The Acidianus two-tailed virus encodes the RIP factor that binds inside the DNA-binding channel of RNAP, inhibiting transcription by occlusion of binding sites for nucleic acid and the transcription initiation factor TFB. Infection with the Sulfolobus Turreted Icosahedral Virus induces the expression of the host factor TFS4, which binds in the RNAP funnel similarly to eukaryotic transcript cleavage factors. However, TFS4 allosterically induces a widening of the DNA-binding channel which disrupts trigger loop and bridge helix motifs. Importantly, the conformational changes induced by TFS4 are closely related to inactivated states of RNAP in other domains of life indicating a deep evolutionary conservation of allosteric RNAP inhibition.
- RNAP Laboratory, Institute for Structural and Molecular Biology, University College London, London, UK.
Organizational Affiliation: