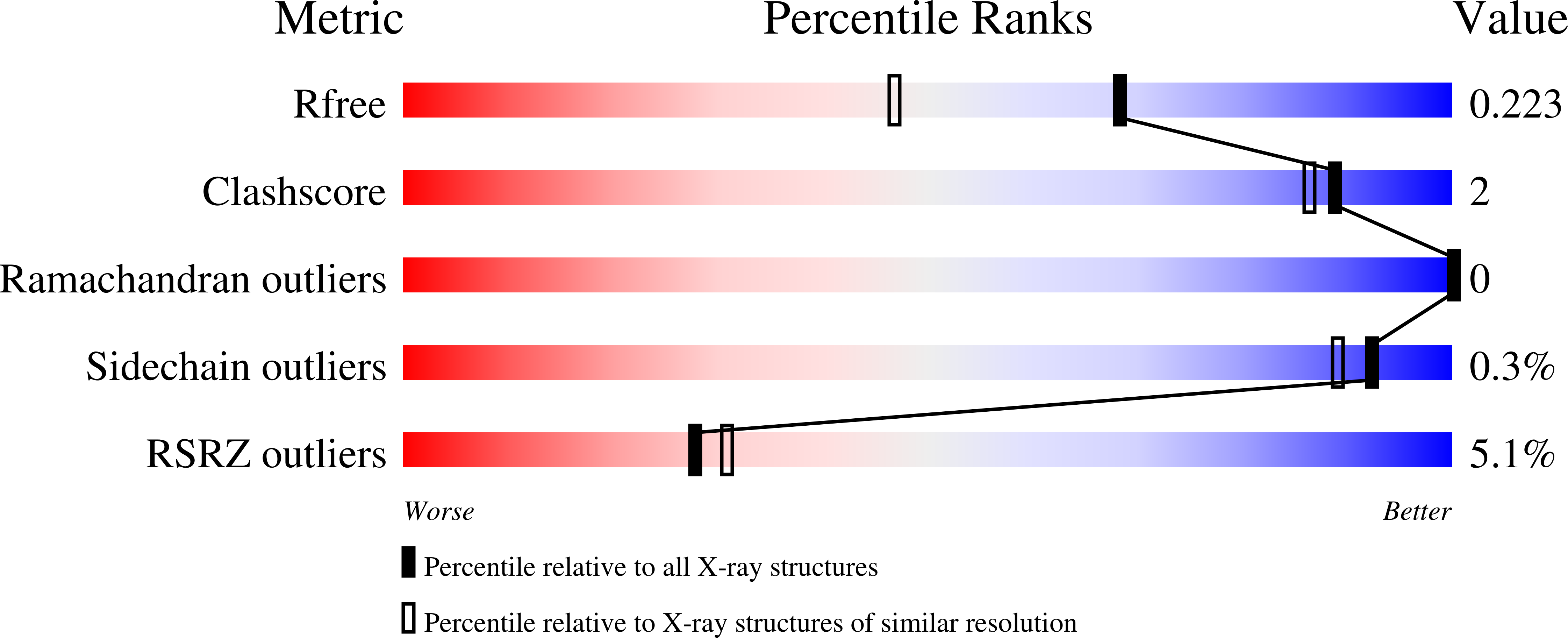
Systematic Engineering of Optimized Autonomous Heavy-Chain Variable Domains.
Nilvebrant, J., Ereno-Orbea, J., Gorelik, M., Julian, M.C., Tessier, P.M., Julien, J.P., Sidhu, S.S.(2021) J Mol Biol 433: 167241-167241
- PubMed: 34508727
- DOI: https://doi.org/10.1016/j.jmb.2021.167241
- Primary Citation of Related Structures:
7OMN, 7OOI - PubMed Abstract:
Autonomous heavy-chain variable (V H ) domains are the smallest functional antibody fragments, and they possess unique features, including small size and convex paratopes, which provide enhanced targeting of concave epitopes that are difficult to access with larger conventional antibodies. However, human V H domains have evolved to fold and function with a light chain partner, and alone, they typically suffer from low stability and high aggregation propensity. Development of autonomous human V H domains, in which aggregation propensity is reduced without compromising antigen recognition, has proven challenging. Here, we used an autonomous human V H domain as a scaffold to construct phage-displayed synthetic libraries in which aspartate was systematically incorporated at different paratope positions. In selections, the library yielded many anti-EphA1 receptor V H domains, which were characterized in detail. Structural analyses of a parental anti-EphA1 V H domain and an improved variant provided insights into the effects of aspartate and other substitutions on preventing aggregation while retaining function. Our naïve libraries and in vitro selection procedures offer a systematic approach to generating highly functional autonomous human V H domains that resist aggregation and could be used for basic research and biomedical applications.
Organizational Affiliation:
Banting and Best Department of Medical Research and Department of Molecular Genetics, The Donnelly Centre, University of Toronto, Toronto, Ontario M5S 3E1, Canada.