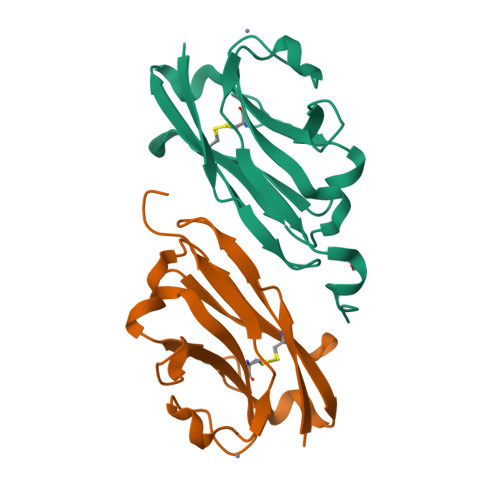
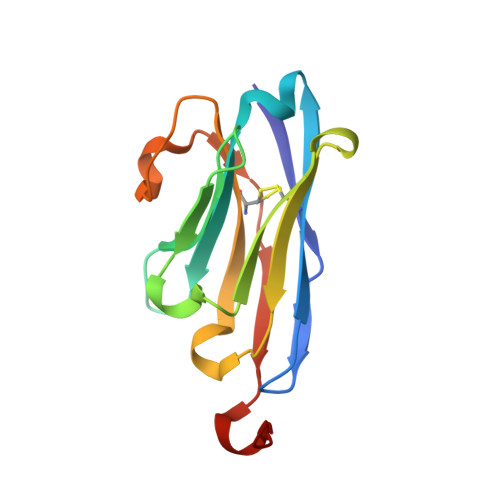
Structural insights into the non-inhibitory mechanism of the anti-EGFR EgB4 nanobody.
Zeronian, M.R., Doulkeridou, S., van Bergen En Henegouwen, P.M.P., Janssen, B.J.C.(2022) BMC Mol Cell Biol 23: 12-12
- PubMed: 35232398
- DOI: https://doi.org/10.1186/s12860-022-00412-x
- Primary Citation of Related Structures:
7OM4, 7OM5 - PubMed Abstract:
The epidermal growth factor receptor (EGFR) is involved in various developmental processes, and alterations of its extracellular segment are associated with several types of cancers, in particular glioblastoma multiforme (GBM). The EGFR extracellular region is therefore a primary target for therapeutic agents, such as monoclonal antibodies and variable domains of heavy chain antibodies (VHH), also called nanobodies. Nanobodies have been previously shown to bind to EGFR, and to inhibit ligand-mediated EGFR activation. Here we present the X-ray crystal structures of the EgB4 nanobody, alone (to 1.48 Å resolution) and bound to the full extracellular EGFR-EGF complex in its active conformation (to 6.0 Å resolution). We show that EgB4 binds to a new epitope located on EGFR domains I and II, and we describe the molecular mechanism by which EgB4 plays a non-inhibitory role in EGFR signaling. This work provides the structural basis for the application of EgB4 as a tool for research, for targeted therapy, or as a biomarker to locate EGFR-associated tumors, all without affecting EGFR activation.
Organizational Affiliation:
Structural Biochemistry, Bijvoet Center for Biomolecular Research, Department of Chemistry, Faculty of Science, Utrecht University, Utrecht, The Netherlands.