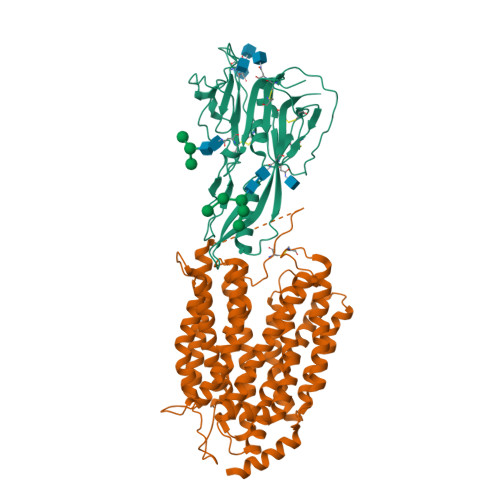
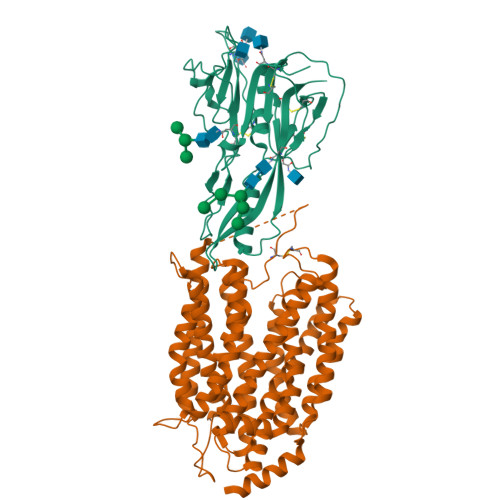
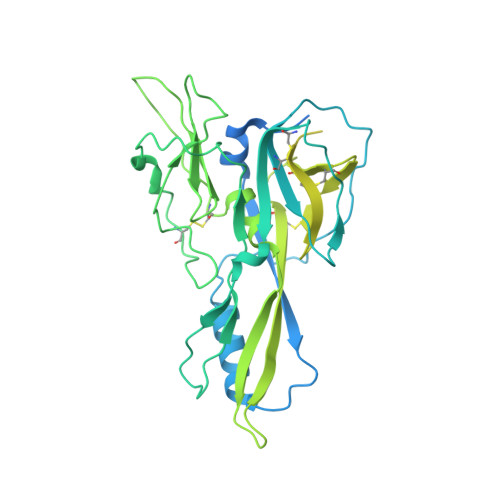
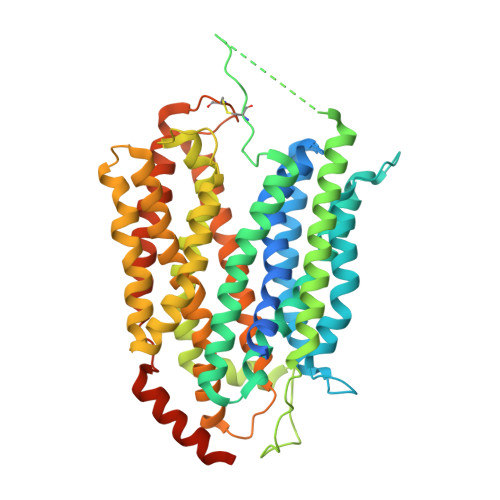
Structural insights into the lysophospholipid brain uptake mechanism and its inhibition by syncytin-2.
Martinez-Molledo, M., Nji, E., Reyes, N.(2022) Nat Struct Mol Biol 29: 604-612
- PubMed: 35710838
- DOI: https://doi.org/10.1038/s41594-022-00786-8
- Primary Citation of Related Structures:
7OIX - PubMed Abstract:
Brain development and function require uptake of essential omega-3 fatty acids in the form of lysophosphatidylcholine via major-facilitator superfamily transporter MFSD2A, a potential pharmaceutical target to modulate blood-brain barrier (BBB) permeability. MFSD2A is also the receptor of endogenous retroviral envelope syncytin-2 (SYNC2) in human placenta, where it mediates cell-cell fusion and formation of the maternal-fetal interface. Here, we report a cryo-electron microscopy structure of the human MFSD2A-SYNC2 complex that reveals a large hydrophobic cavity in the transporter C-terminal domain to occlude long aliphatic chains. The transporter architecture suggests an alternating-access transport mechanism for lipid substrates in mammalian MFS transporters. SYNC2 establishes an extensive binding interface with MFSD2A, and a SYNC2-soluble fragment acts as a long-sought-after inhibitor of MFSD2A transport. Our work uncovers molecular mechanisms important to brain and placenta development and function, and SYNC2-mediated inhibition of MFSD2A transport suggests strategies to aid delivery of therapeutic macromolecules across the BBB.
Organizational Affiliation:
Membrane Protein Mechanisms Group, European Institute of Chemistry and Biology, University of Bordeaux, CNRS-UMR5234, Bordeaux, France.