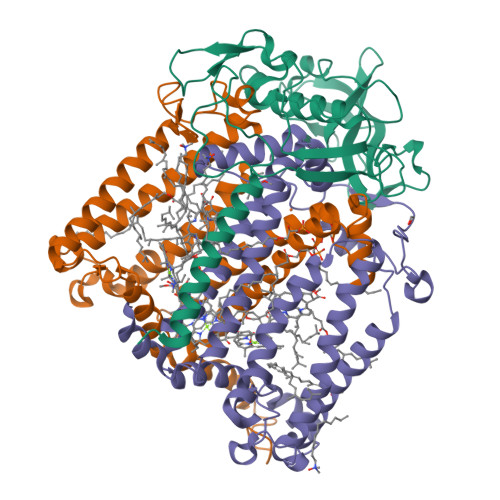
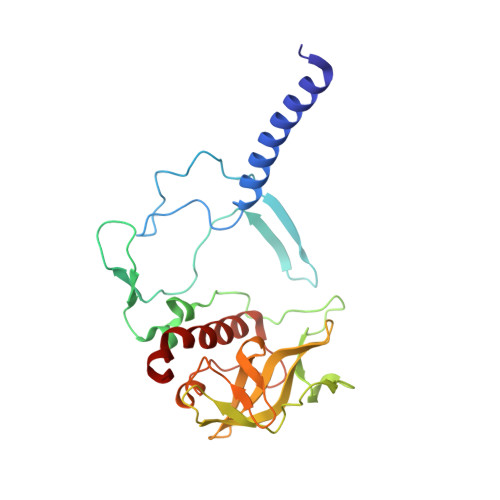
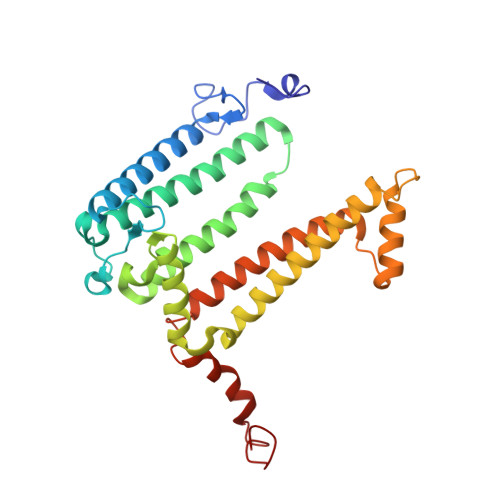
X-ray structure of the Rhodobacter sphaeroides reaction center with an M197 Phe→His substitution clarifies the properties of the mutant complex.
Selikhanov, G., Fufina, T., Guenther, S., Meents, A., Gabdulkhakov, A., Vasilieva, L.(2022) IUCrJ 9: 261-271
- PubMed: 35371503
- DOI: https://doi.org/10.1107/S2052252521013178
- Primary Citation of Related Structures:
7OD5, 7P2C - PubMed Abstract:
The first steps of the global process of photosynthesis take place in specialized membrane pigment-protein complexes called photosynthetic reaction centers (RCs). The RC of the photosynthetic purple bacterium Rhodobacter sphaeroides , a relatively simple analog of the more complexly organized photosystem II in plants, algae and cyanobacteria, serves as a convenient model for studying pigment-protein interactions that affect photochemical processes. In bacterial RCs the bacteriochlorophyll (BChl) dimer P serves as the primary electron donor, and its redox potential is a critical factor in the efficient functioning of the RC. It has previously been shown that the replacement of Phe M197 by His strongly affects the oxidation potential of P ( E m P/P + ), increasing its value by 125 mV, as well as increasing the thermal stability of RC and its stability in response to external pressure. The crystal structures of F(M197)H RC at high resolution obtained using various techniques presented in this report clarify the optical and electrochemical properties of the primary electron donor and the increased resistance of the mutant complex to denaturation. The electron-density maps are consistent with the donation of a hydrogen bond from the imidazole group of His M197 to the C2-acetyl carbonyl group of BChl P B . The formation of this hydrogen bond leads to a considerable out-of-plane rotation of the acetyl carbonyl group and results in a 1.2 Å shift of the O atom of this group relative to the wild-type structure. Besides, the distance between BChl P A and P B in the area of pyrrole ring I was found to be increased by up to 0.17 Å. These structural changes are discussed in association with the spectral properties of BChl dimer P. The electron-density maps strongly suggest that the imidazole group of His M197 accepts another hydrogen bond from the nearest water molecule, which in turn appears to form two more hydrogen bonds to Asn M195 and Asp L155. As a result of the F(M197)H mutation, BChl P B finds itself connected to the extensive hydrogen-bonding network that pre-existed in wild-type RC. Dissimilarities in the two hydrogen-bonding networks near the M197 and L168 sites may account for the different changes of the E m P/P + in F(M197)H and H(L168)F RCs. The involvement of His M197 in the hydrogen-bonding network also appears to be related to stabilization of the F(M197)H RC structure. Analysis of the experimental data presented here and of the data available in the literature points to the fact that the hydrogen-bonding networks in the vicinity of BChl dimer P may play an important role in fine-tuning the redox properties of the primary electron donor.
Organizational Affiliation:
Group of Structural Studies of Macromolecular Complexes, Institute of Protein Research, Russian Academy of Sciences, Institutskaya 4, Pushchino 142290, Moscow Region, Russian Federation.