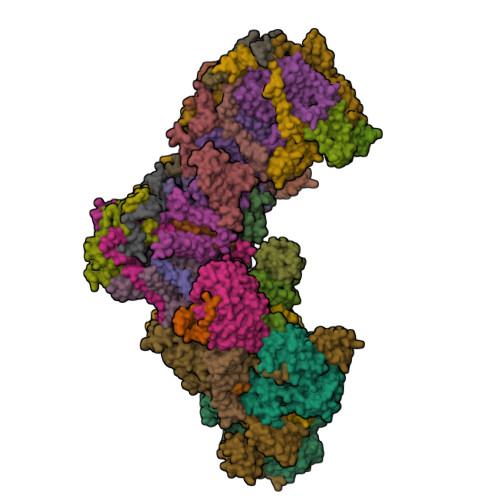
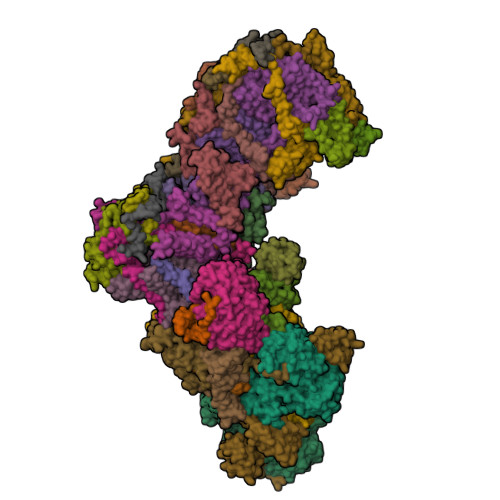
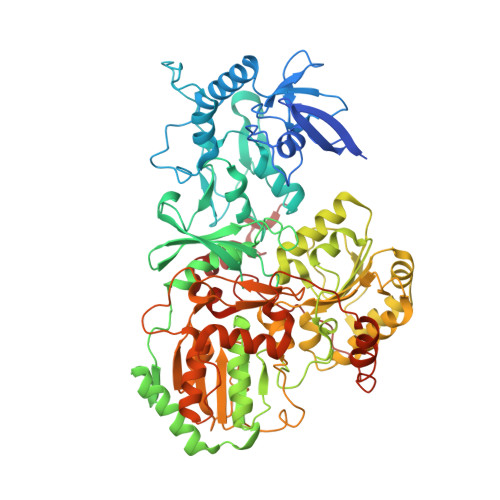
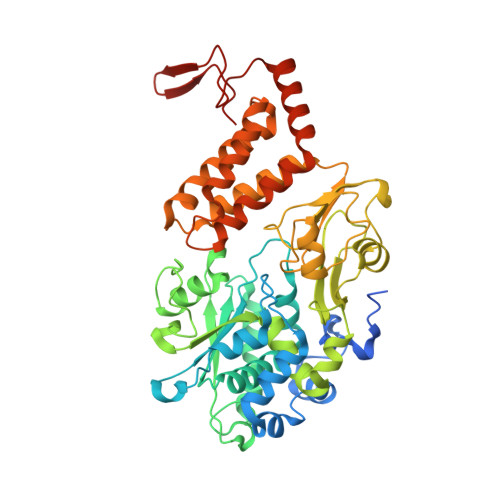
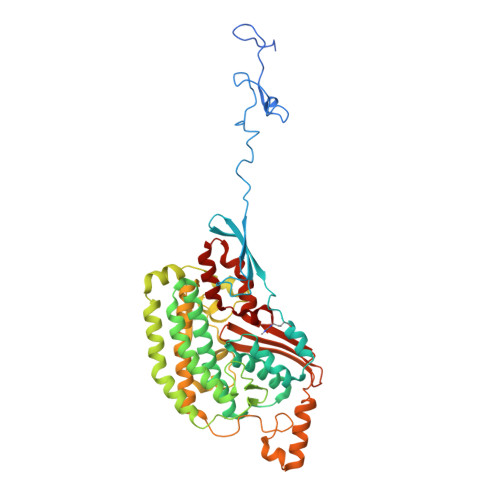
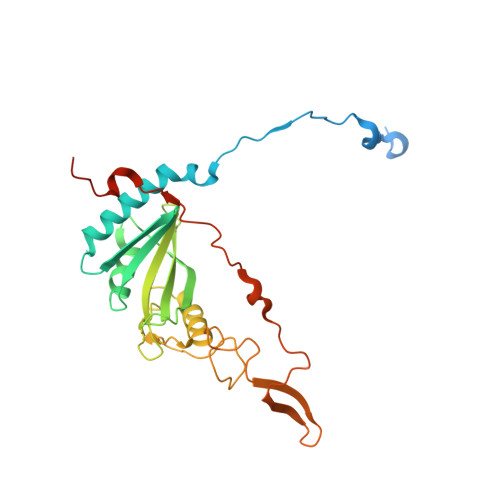
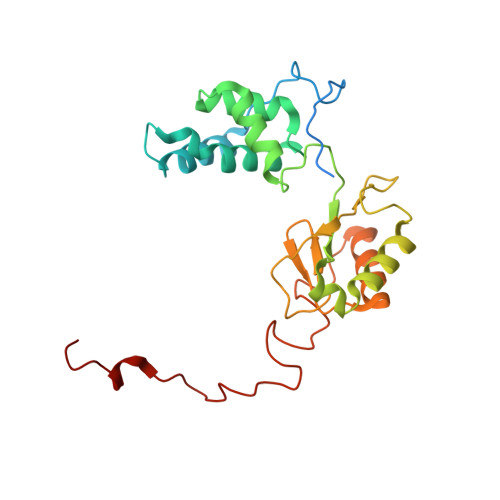
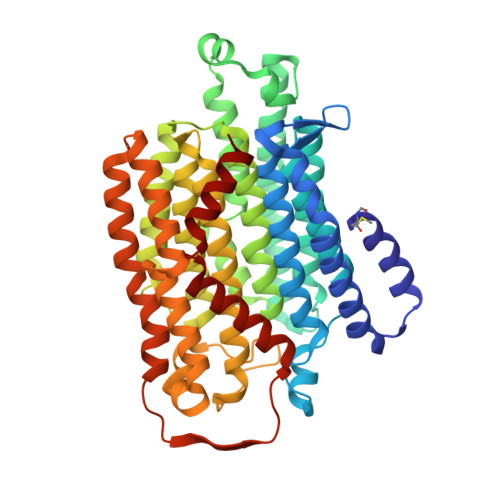
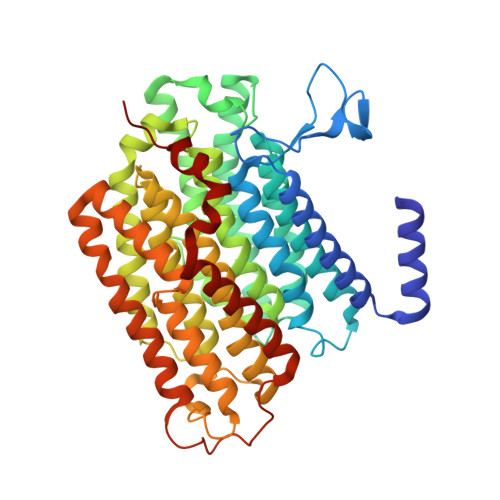
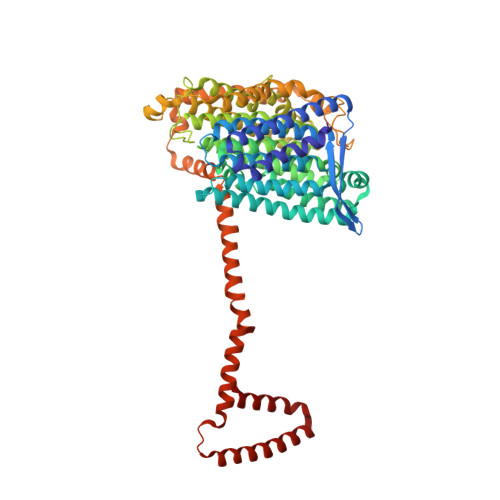
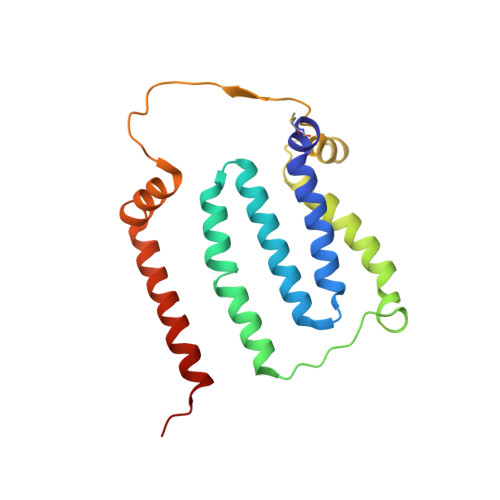
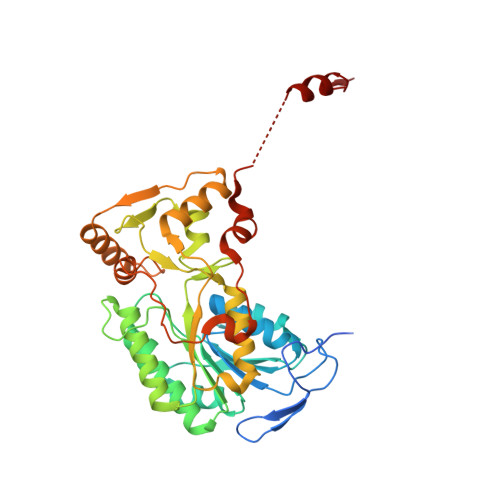
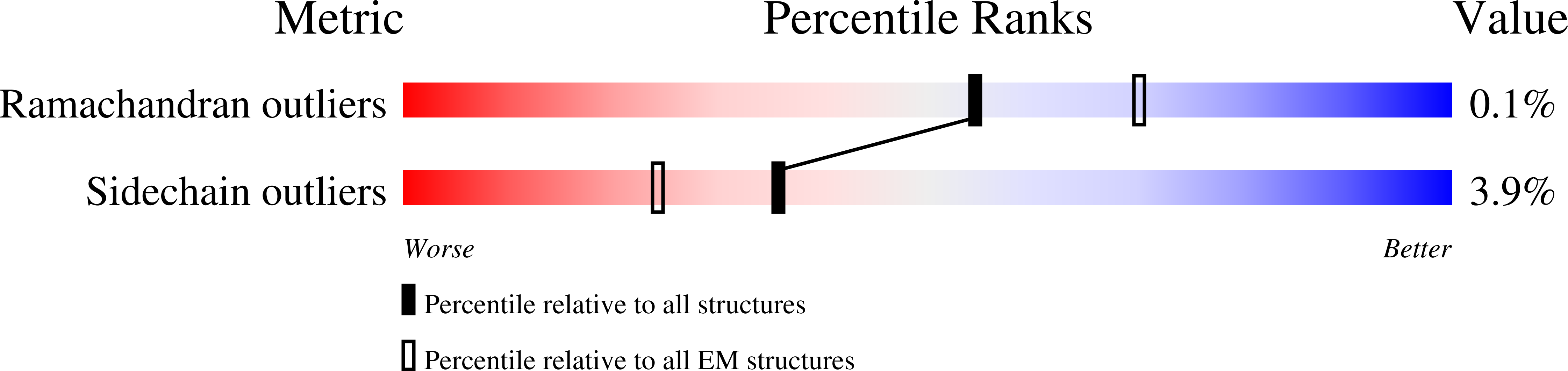High-resolution structure and dynamics of mitochondrial complex I-Insights into the proton pumping mechanism.
Parey, K., Lasham, J., Mills, D.J., Djurabekova, A., Haapanen, O., Yoga, E.G., Xie, H., Kuhlbrandt, W., Sharma, V., Vonck, J., Zickermann, V.(2021) Sci Adv 7: eabj3221-eabj3221
- PubMed: 34767441
- DOI: https://doi.org/10.1126/sciadv.abj3221
- Primary Citation of Related Structures:
7O6Y, 7O71 - PubMed Abstract:
Mitochondrial NADH:ubiquinone oxidoreductase (complex I) is a 1-MDa membrane protein complex with a central role in energy metabolism. Redox-driven proton translocation by complex I contributes substantially to the proton motive force that drives ATP synthase. Several structures of complex I from bacteria and mitochondria have been determined, but its catalytic mechanism has remained controversial. We here present the cryo-EM structure of complex I from Yarrowia lipolytica at 2.1-Å resolution, which reveals the positions of more than 1600 protein-bound water molecules, of which ~100 are located in putative proton translocation pathways. Another structure of the same complex under steady-state activity conditions at 3.4-Å resolution indicates conformational transitions that we associate with proton injection into the central hydrophilic axis. By combining high-resolution structural data with site-directed mutagenesis and large-scale molecular dynamic simulations, we define details of the proton translocation pathways and offer insights into the redox-coupled proton pumping mechanism of complex I.
Organizational Affiliation:
Institute of Biochemistry II, University Hospital, Goethe University, 60590 Frankfurt am Main, Germany.