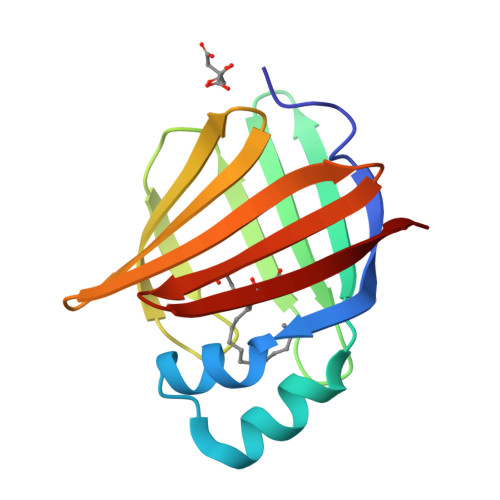
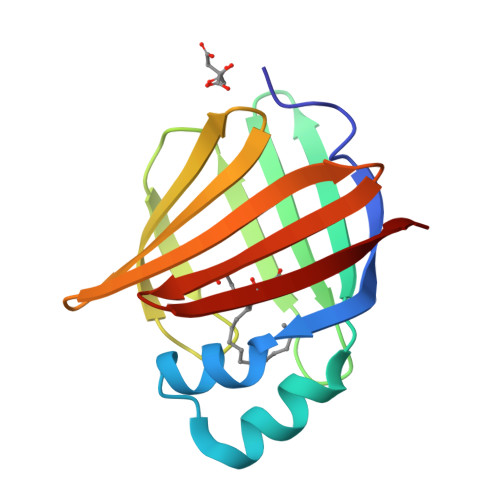
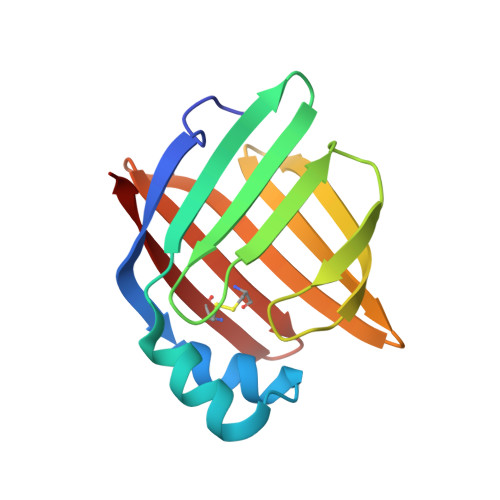
Human myelin protein P2: from crystallography to time-lapse membrane imaging and neuropathy-associated variants.
Uusitalo, M., Klenow, M.B., Laulumaa, S., Blakeley, M.P., Simonsen, A.C., Ruskamo, S., Kursula, P.(2021) FEBS J 288: 6716-6735
- PubMed: 34138518
- DOI: https://doi.org/10.1111/febs.16079
- Primary Citation of Related Structures:
7NRW, 7NSR, 7NTP, 7O60 - PubMed Abstract:
Peripheral myelin protein 2 (P2) is a fatty acid-binding protein expressed in vertebrate peripheral nervous system myelin, as well as in human astrocytes. Suggested functions of P2 include membrane stacking and lipid transport. Mutations in the PMP2 gene, encoding P2, are associated with Charcot-Marie-Tooth disease (CMT). Recent studies have revealed three novel PMP2 mutations in CMT patients. To shed light on the structure and function of these P2 variants, we used X-ray and neutron crystallography, small-angle X-ray scattering, circular dichroism spectroscopy, computer simulations and lipid binding assays. The crystal and solution structures of the I50del, M114T and V115A variants of P2 showed minor differences to the wild-type protein, whereas their thermal stability was reduced. Vesicle aggregation assays revealed no change in membrane stacking characteristics, while the variants showed altered fatty acid binding. Time-lapse imaging of lipid bilayers indicated formation of double-membrane structures induced by P2, which could be related to its function in stacking of two myelin membrane surfaces in vivo. In order to better understand the links between structure, dynamics and function, the crystal structure of perdeuterated P2 was refined from room temperature data using neutrons and X-rays, and the results were compared to simulations and cryocooled crystal structures. Our data indicate similar properties for all known human P2 CMT variants; while crystal structures are nearly identical, thermal stability and function of CMT variants are impaired. Our data provide new insights into the structure-function relationships and dynamics of P2 in health and disease.
Organizational Affiliation:
Faculty of Biochemistry and Molecular Medicine & Biocenter Oulu, University of Oulu, Finland.