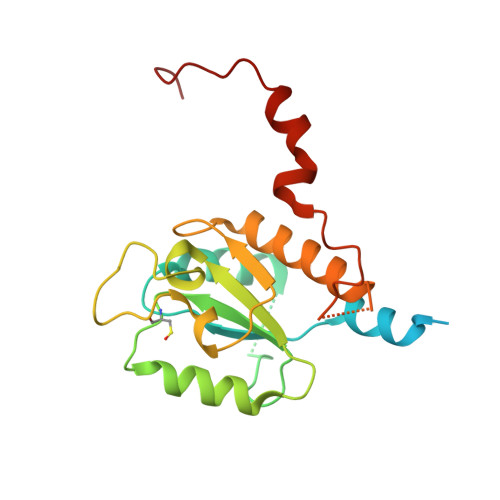
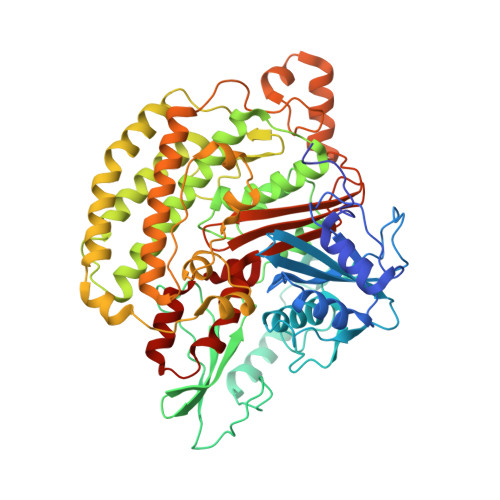
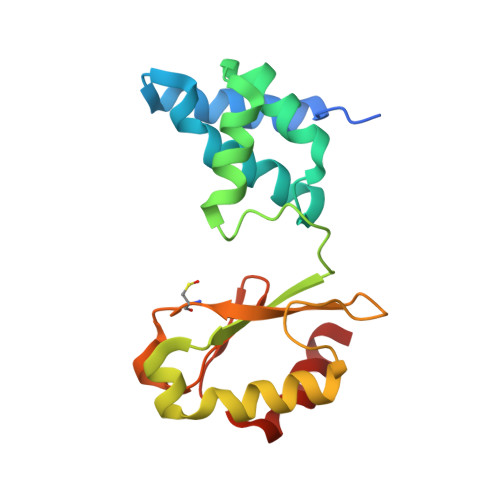
Structure of Escherichia coli respiratory complex I reconstituted into lipid nanodiscs reveals an uncoupled conformation.
Kolata, P., Efremov, R.G.(2021) Elife 10
- PubMed: 34308841
- DOI: https://doi.org/10.7554/eLife.68710
- Primary Citation of Related Structures:
7NYH, 7NYR, 7NYU, 7NYV, 7NZ1 - PubMed Abstract:
Respiratory complex I is a multi-subunit membrane protein complex that reversibly couples NADH oxidation and ubiquinone reduction with proton translocation against transmembrane potential. Complex I from Escherichia coli is among the best functionally characterized complexes, but its structure remains unknown, hindering further studies to understand the enzyme coupling mechanism. Here, we describe the single particle cryo-electron microscopy (cryo-EM) structure of the entire catalytically active E. coli complex I reconstituted into lipid nanodiscs. The structure of this mesophilic bacterial complex I displays highly dynamic connection between the peripheral and membrane domains. The peripheral domain assembly is stabilized by unique terminal extensions and an insertion loop. The membrane domain structure reveals novel dynamic features. Unusual conformation of the conserved interface between the peripheral and membrane domains suggests an uncoupled conformation of the complex. Considering constraints imposed by the structural data, we suggest a new simple hypothetical coupling mechanism for the molecular machine.
Organizational Affiliation:
Center for Structural Biology, Vlaams Instituut voor Biotechnologie, Brussels, Belgium.