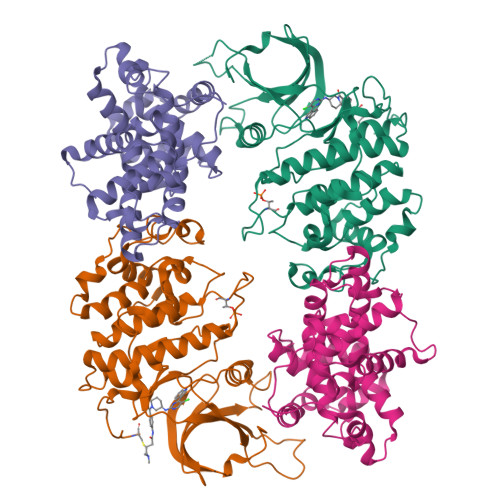
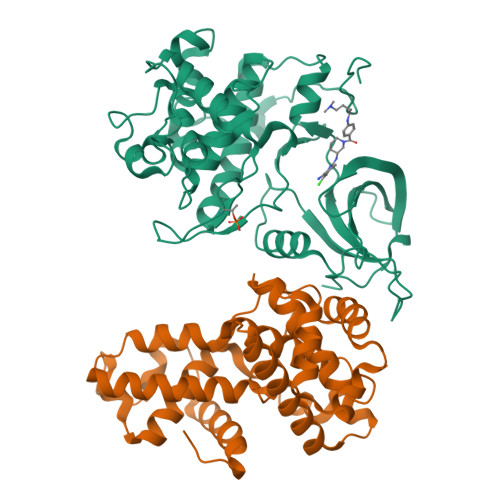
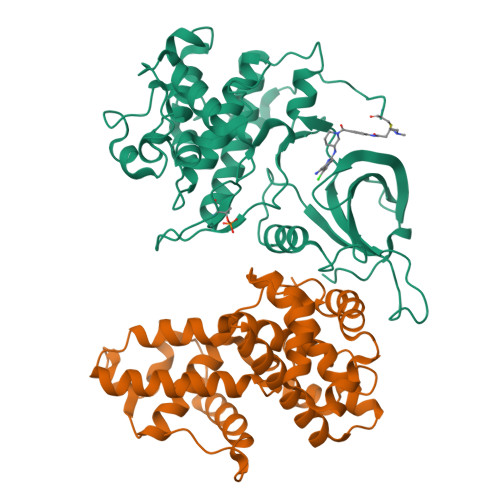
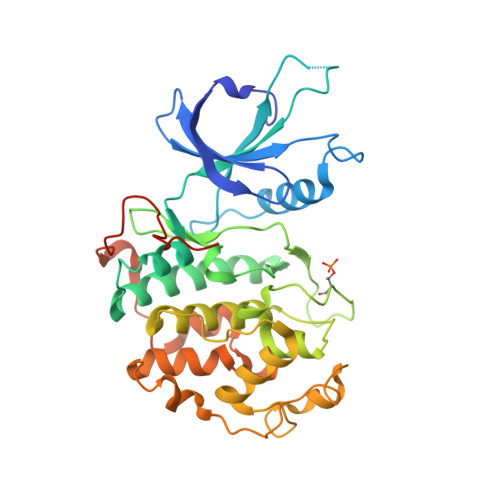
Structure-activity relationship study of THZ531 derivatives enables the discovery of BSJ-01-175 as a dual CDK12/13 covalent inhibitor with efficacy in Ewing sarcoma.
Jiang, B., Jiang, J., Kaltheuner, I.H., Iniguez, A.B., Anand, K., Ferguson, F.M., Ficarro, S.B., Seong, B.K.A., Greifenberg, A.K., Dust, S., Kwiatkowski, N.P., Marto, J.A., Stegmaier, K., Zhang, T., Geyer, M., Gray, N.S.(2021) Eur J Med Chem 221: 113481-113481
- PubMed: 33945934
- DOI: https://doi.org/10.1016/j.ejmech.2021.113481
- Primary Citation of Related Structures:
7NXJ, 7NXK - PubMed Abstract:
Development of inhibitors targeting CDK12/13 is of increasing interest as a potential therapy for cancers as these compounds inhibit transcription of DNA damage response (DDR) genes. We previously described THZ531, a covalent inhibitor with selectivity for CDK12/13. In order to elucidate structure-activity relationship (SAR), we have undertaken a medicinal chemistry campaign and established a focused library of THZ531 analogs. Among these analogs, BSJ-01-175 demonstrates exquisite selectivity, potent inhibition of RNA polymerase II phosphorylation, and downregulation of CDK12-targeted genes in cancer cells. A 3.0 Å co-crystal structure with CDK12/CycK provides a structural rational for selective targeting of Cys1039 located in a C-terminal extension from the kinase domain. With moderate pharmacokinetic properties, BSJ-01-175 exhibits efficacy against an Ewing sarcoma tumor growth in a patient-derived xenograft (PDX) mouse model following 10 mg/kg once a day, intraperitoneal administration. Taken together, BSJ-01-175 represents the first selective CDK12/13 covalent inhibitor with in vivo efficacy reported to date.
Organizational Affiliation:
Department of Cancer Biology, Dana-Farber Cancer Institute, Harvard Medical School, Boston, MA, 02115, USA; Department of Biological Chemistry and Molecular Pharmacology, Harvard Medical School, Boston, MA, 02115, USA.