Small-molecule modulators of TRMT2A decrease PolyQ aggregation and PolyQ-induced cell death.
Margreiter, M.A., Witzenberger, M., Wasser, Y., Davydova, E., Janowski, R., Metz, J., Habib, P., Sahnoun, S.E.M., Sobisch, C., Poma, B., Palomino-Hernandez, O., Wagner, M., Carell, T., Jon Shah, N., Schulz, J.B., Niessing, D., Voigt, A., Rossetti, G.(2022) Comput Struct Biotechnol J 20: 443-458
- PubMed: 35070167
- DOI: https://doi.org/10.1016/j.csbj.2021.12.029
- Primary Citation of Related Structures:
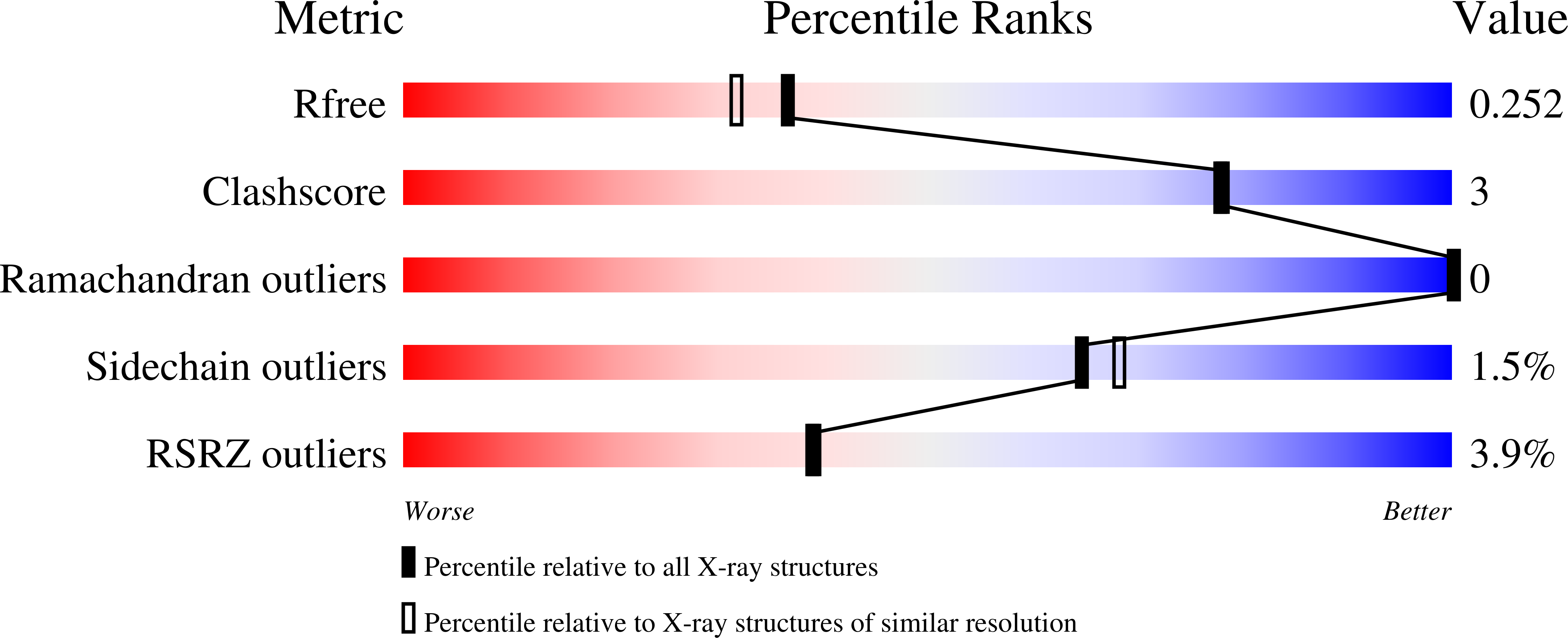
7NTN, 7NTO - PubMed Abstract:
Polyglutamine (polyQ) diseases are characterized by an expansion of cytosine-adenine-guanine (CAG) trinucleotide repeats encoding for an uninterrupted prolonged polyQ tract. We previously identified TRMT2A as a strong modifier of polyQ-induced toxicity in an unbiased large-scale screen in Drosophila melanogaster . This work aimed at identifying and validating pharmacological TRMT2A inhibitors as treatment opportunities for polyQ diseases in humans. Computer-aided drug discovery was implemented to identify human TRMT2A inhibitors. Additionally, the crystal structure of one protein domain, the RNA recognition motif (RRM), was determined, and Biacore experiments with the RRM were performed. The identified molecules were validated for their potency to reduce polyQ aggregation and polyQ-induced cell death in human HEK293T cells and patient derived fibroblasts. Our work provides a first step towards pharmacological inhibition of this enzyme and indicates TRMT2A as a viable drug target for polyQ diseases.
Organizational Affiliation:
Institute of Neuroscience and Medicine (INM-9), Forschungszentrum Juelich GmbH, Germany.