Structural basis of coronavirus E protein interactions with human PALS1 PDZ domain.
Javorsky, A., Humbert, P.O., Kvansakul, M.(2021) Commun Biol 4: 724-724
- PubMed: 34117354
- DOI: https://doi.org/10.1038/s42003-021-02250-7
- Primary Citation of Related Structures:
7NTJ, 7NTK - PubMed Abstract:
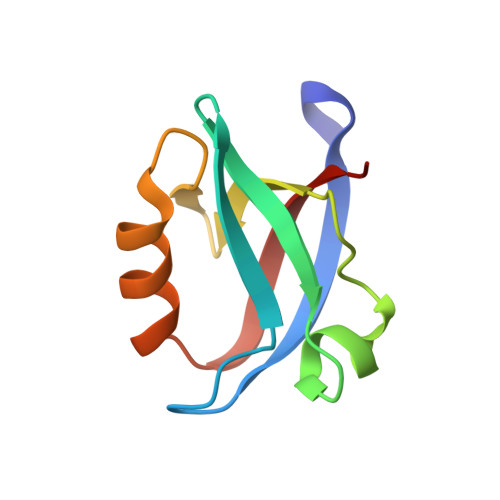
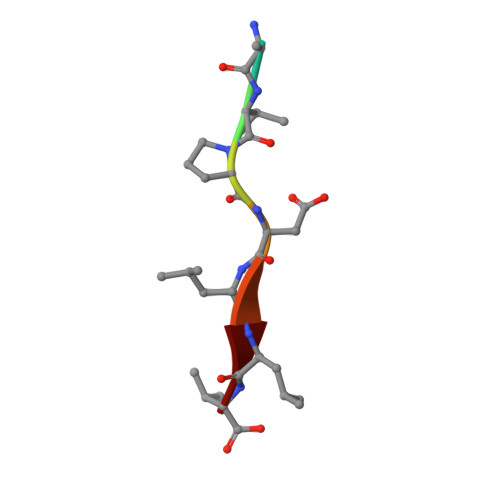
SARS-CoV-2 infection leads to coronavirus disease 2019 (COVID-19), which is associated with severe and life-threatening pneumonia and respiratory failure. However, the molecular basis of these symptoms remains unclear. SARS-CoV-1 E protein interferes with control of cell polarity and cell-cell junction integrity in human epithelial cells by binding to the PALS1 PDZ domain, a key component of the Crumbs polarity complex. We show that C-terminal PDZ binding motifs of SARS-CoV-1 and SARS-CoV-2 E proteins bind the PALS1 PDZ domain with 29.6 and 22.8 μM affinity, whereas the related sequence from MERS-CoV did not bind. We then determined crystal structures of PALS1 PDZ domain bound to both SARS-CoV-1 and SARS-CoV-2 E protein PDZ binding motifs. Our findings establish the structural basis for SARS-CoV-1/2 mediated subversion of Crumbs polarity signalling and serve as a platform for the development of small molecule inhibitors to suppress SARS-CoV-1/2 mediated disruption of polarity signalling in epithelial cells.
Organizational Affiliation:
Department of Biochemistry & Genetics, La Trobe Institute for Molecular Science, La Trobe University, Melbourne, Vic, Australia.