Structure and dynamics of a mycobacterial type VII secretion system.
Bunduc, C.M., Fahrenkamp, D., Wald, J., Ummels, R., Bitter, W., Houben, E.N.G., Marlovits, T.C.(2021) Nature 593: 445-448
- PubMed: 33981042
- DOI: https://doi.org/10.1038/s41586-021-03517-z
- Primary Citation of Related Structures:
7NP7, 7NPR, 7NPS, 7NPT, 7NPU, 7NPV - PubMed Abstract:
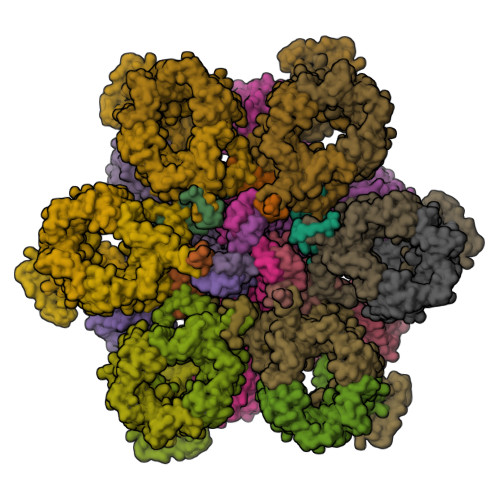
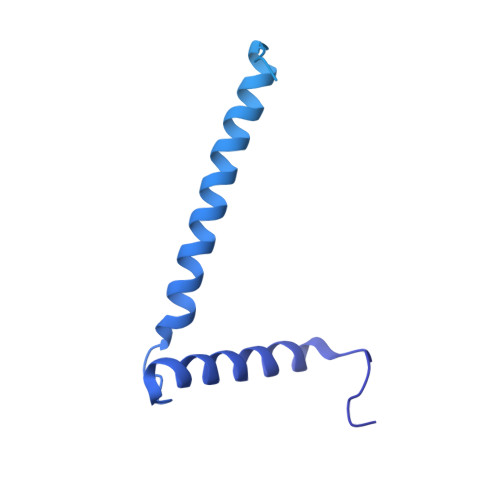
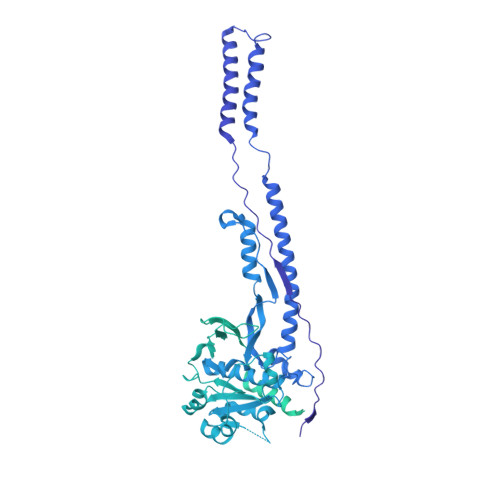
Mycobacterium tuberculosis is the cause of one of the most important infectious diseases in humans, which leads to 1.4 million deaths every year 1 . Specialized protein transport systems-known as type VII secretion systems (T7SSs)-are central to the virulence of this pathogen, and are also crucial for nutrient and metabolite transport across the mycobacterial cell envelope 2,3 . Here we present the structure of an intact T7SS inner-membrane complex of M. tuberculosis. We show how the 2.32-MDa ESX-5 assembly, which contains 165 transmembrane helices, is restructured and stabilized as a trimer of dimers by the MycP 5 protease. A trimer of MycP 5 caps a central periplasmic dome-like chamber that is formed by three EccB 5 dimers, with the proteolytic sites of MycP 5 facing towards the cavity. This chamber suggests a central secretion and processing conduit. Complexes without MycP 5 show disruption of the EccB 5 periplasmic assembly and increased flexibility, which highlights the importance of MycP 5 for complex integrity. Beneath the EccB 5 -MycP 5 chamber, dimers of the EccC 5 ATPase assemble into three bundles of four transmembrane helices each, which together seal the potential central secretion channel. Individual cytoplasmic EccC 5 domains adopt two distinctive conformations that probably reflect different secretion states. Our work suggests a previously undescribed mechanism of protein transport and provides a structural scaffold to aid in the development of drugs against this major human pathogen.
Organizational Affiliation:
Centre for Structural Systems Biology, Hamburg, Germany.