The molecular basis of regulation of bacterial capsule assembly by Wzc.
Yang, Y., Liu, J., Clarke, B.R., Seidel, L., Bolla, J.R., Ward, P.N., Zhang, P., Robinson, C.V., Whitfield, C., Naismith, J.H.(2021) Nat Commun 12: 4349-4349
- PubMed: 34272394
- DOI: https://doi.org/10.1038/s41467-021-24652-1
- Primary Citation of Related Structures:
7NHR, 7NHS, 7NI2, 7NIB, 7NIH, 7NII - PubMed Abstract:
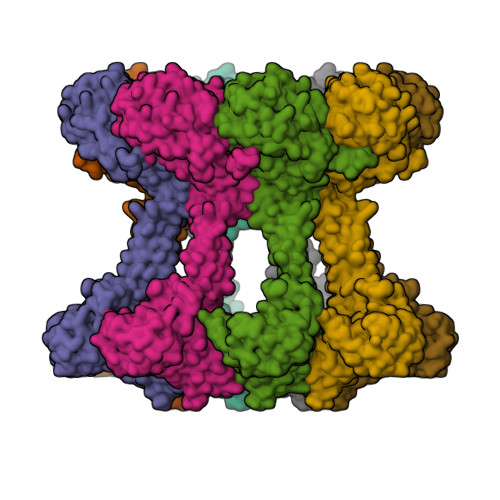
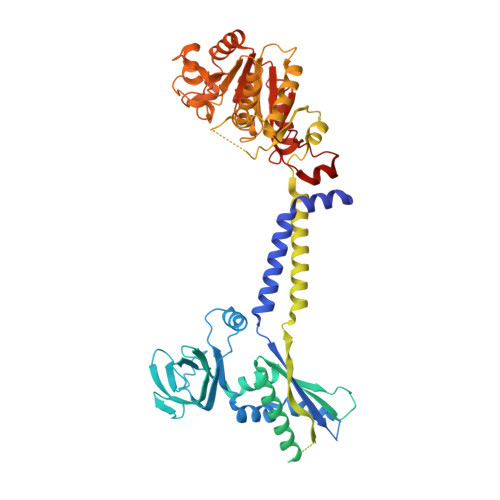
Bacterial extracellular polysaccharides (EPSs) play critical roles in virulence. Many bacteria assemble EPSs via a multi-protein "Wzx-Wzy" system, involving glycan polymerization at the outer face of the cytoplasmic/inner membrane. Gram-negative species couple polymerization with translocation across the periplasm and outer membrane and the master regulator of the system is the tyrosine autokinase, Wzc. This near atomic cryo-EM structure of dephosphorylated Wzc from E. coli shows an octameric assembly with a large central cavity formed by transmembrane helices. The tyrosine autokinase domain forms the cytoplasm region, while the periplasmic region contains small folded motifs and helical bundles. The helical bundles are essential for function, most likely through interaction with the outer membrane translocon, Wza. Autophosphorylation of the tyrosine-rich C-terminus of Wzc results in disassembly of the octamer into multiply phosphorylated monomers. We propose that the cycling between phosphorylated monomer and dephosphorylated octamer regulates glycan polymerization and translocation.
Organizational Affiliation:
Rosalind Franklin Institute, Harwell Campus, Harwell, UK.