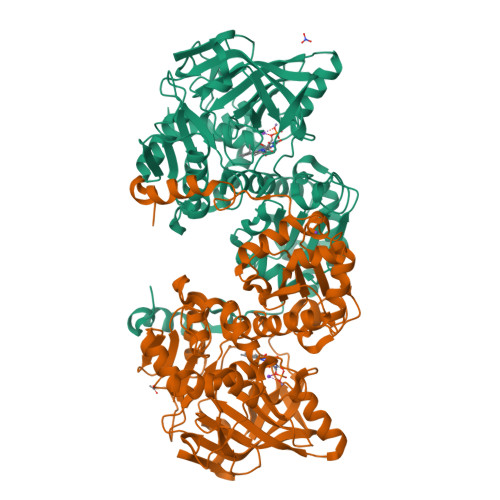
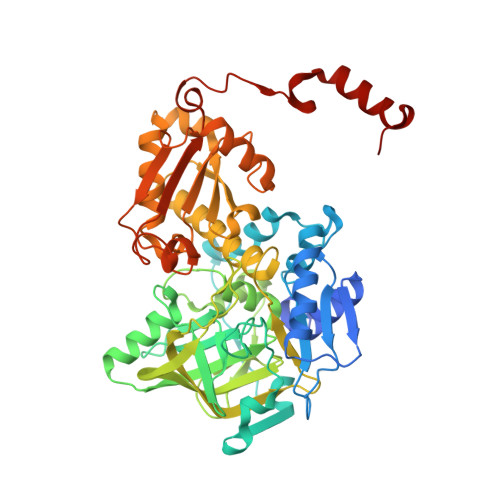
Directed evolution of prenylated FMN-dependent Fdc supports efficient in vivo isobutene production.
Saaret, A., Villiers, B., Stricher, F., Anissimova, M., Cadillon, M., Spiess, R., Hay, S., Leys, D.(2021) Nat Commun 12: 5300-5300
- PubMed: 34489427
- DOI: https://doi.org/10.1038/s41467-021-25598-0
- Primary Citation of Related Structures:
7NEY, 7NF0, 7NF1, 7NF2, 7NF3, 7NF4 - PubMed Abstract:
Isobutene is a high value gaseous alkene used as fuel additive and a chemical building block. As an alternative to fossil fuel derived isobutene, we here develop a modified mevalonate pathway for the production of isobutene from glucose in vivo. The final step in the pathway consists of the decarboxylation of 3-methylcrotonic acid, catalysed by an evolved ferulic acid decarboxylase (Fdc) enzyme. Fdc belongs to the prFMN-dependent UbiD enzyme family that catalyses reversible decarboxylation of (hetero)aromatic acids or acrylic acids with extended conjugation. Following a screen of an Fdc library for inherent 3-methylcrotonic acid decarboxylase activity, directed evolution yields variants with up to an 80-fold increase in activity. Crystal structures of the evolved variants reveal that changes in the substrate binding pocket are responsible for increased selectivity. Solution and computational studies suggest that isobutene cycloelimination is rate limiting and strictly dependent on presence of the 3-methyl group.
Organizational Affiliation:
Department of Chemistry, Manchester Institute for Biotechnology, University of Manchester, Manchester, UK.