Investigating molecular mechanisms of 2A-stimulated ribosomal pausing and frameshifting in Theilovirus.
Hill, C.H., Cook, G.M., Napthine, S., Kibe, A., Brown, K., Caliskan, N., Firth, A.E., Graham, S.C., Brierley, I.(2021) Nucleic Acids Res 49: 11938-11958
- PubMed: 34751406
- DOI: https://doi.org/10.1093/nar/gkab969
- Primary Citation of Related Structures:
7NBV - PubMed Abstract:
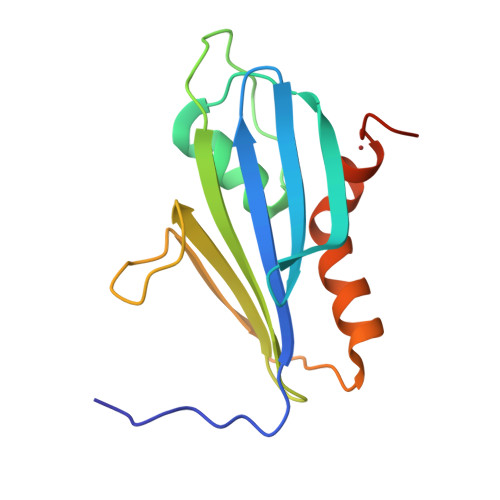
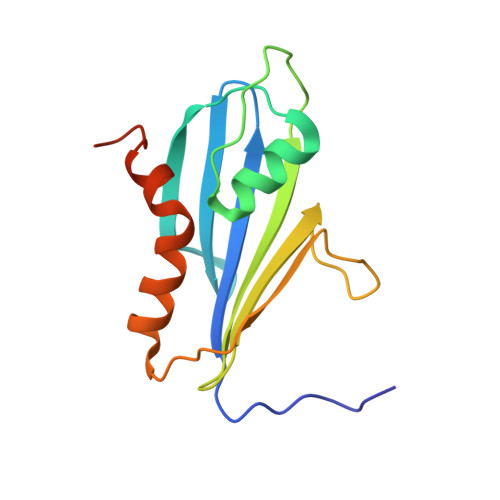
The 2A protein of Theiler's murine encephalomyelitis virus (TMEV) acts as a switch to stimulate programmed -1 ribosomal frameshifting (PRF) during infection. Here, we present the X-ray crystal structure of TMEV 2A and define how it recognises the stimulatory RNA element. We demonstrate a critical role for bases upstream of the originally predicted stem-loop, providing evidence for a pseudoknot-like conformation and suggesting that the recognition of this pseudoknot by beta-shell proteins is a conserved feature in cardioviruses. Through examination of PRF in TMEV-infected cells by ribosome profiling, we identify a series of ribosomal pauses around the site of PRF induced by the 2A-pseudoknot complex. Careful normalisation of ribosomal profiling data with a 2A knockout virus facilitated the identification, through disome analysis, of ribosome stacking at the TMEV frameshifting signal. These experiments provide unparalleled detail of the molecular mechanisms underpinning Theilovirus protein-stimulated frameshifting.
Organizational Affiliation:
Division of Virology, Department of Pathology, University of Cambridge, Tennis Court Road, Cambridge CB2 1QP, UK.