Identification of unprecedented ATP-competitive choline kinase inhibitors.
Quartieri, F., Nesi, M., Avanzi, N.R., Borghi, D., Casale, E., Corti, E., Cucchi, U., Donati, D., Fasolini, M., Felder, E.R., Galvani, A., Giorgini, M.L., Lomolino, A., Menichincheri, M., Orrenius, C., Perrera, C., Re Depaolini, S., Riccardi-Sirtori, F., Salsi, E., Isacchi, A., Gnocchi, P.(2021) Bioorg Med Chem Lett 51: 128310-128310
- PubMed: 34416377
- DOI: https://doi.org/10.1016/j.bmcl.2021.128310
- Primary Citation of Related Structures:
7NB1, 7NB2, 7NB3 - PubMed Abstract:
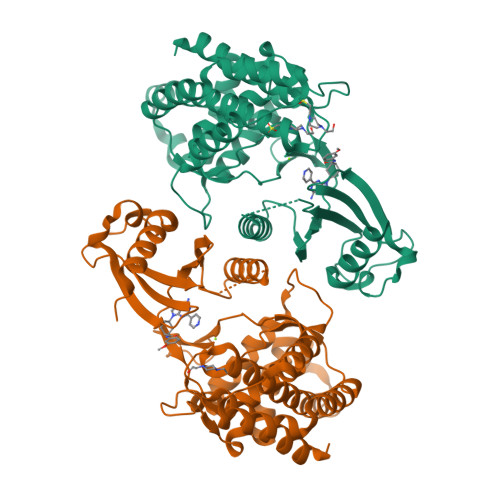
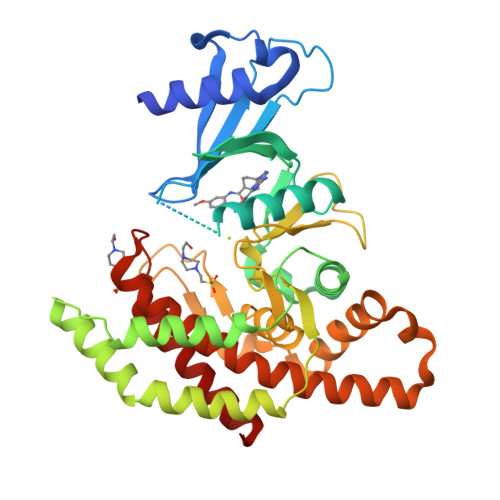
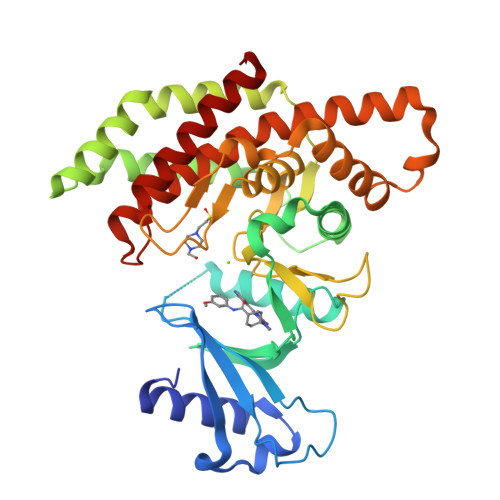
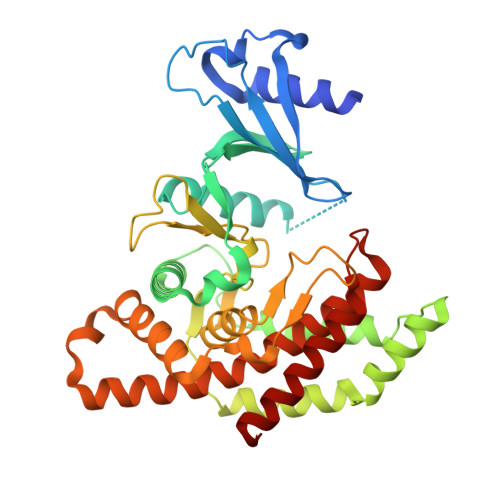
In this article we describe the identification of unprecedented ATP-competitive ChoKα inhibitors starting from initial hit NMS-P830 that binds to ChoKα in an ATP concentration-dependent manner. This result is confirmed by the co-crystal structure of NMS-P830 in complex with Δ75-ChoKα. NMS-P830 is able to inhibit ChoKα in cells resulting in the reduction of intracellular phosphocholine formation. A structure-based medicinal chemistry program resulted in the identification of selective compounds that have good biochemical activity, solubility and metabolic stability and are suitable for further optimization. The ChoKα inhibitors disclosed in this article demonstrate for the first time the possibility to inhibit ChoKα with ATP-competitive compounds.
Organizational Affiliation:
Nerviano Medical Sciences Srl, Viale Pasteur 10, 20014 Nerviano (MI), Italy. Electronic address: francesca.quartieri@nervianoms.com.