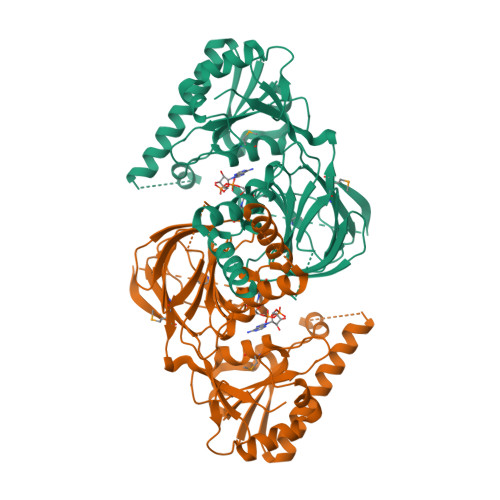
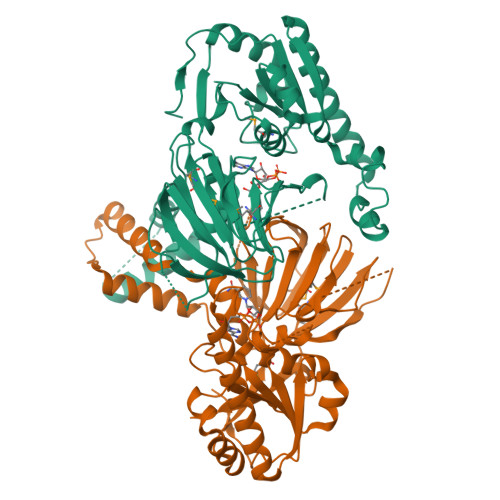
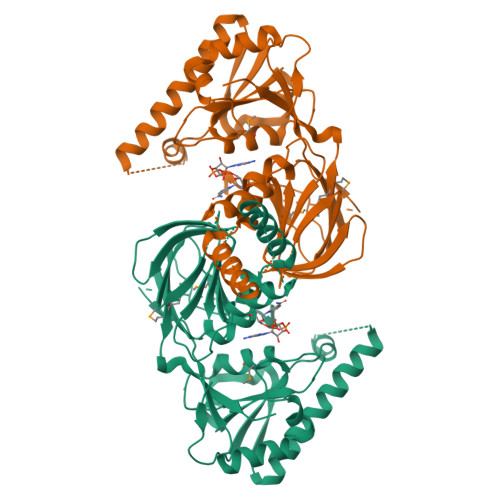
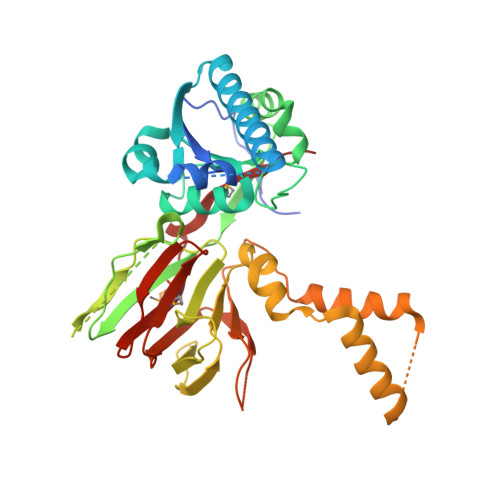
Structure of human NADK2 reveals atypical assembly and regulation of NAD kinases from animal mitochondria.
Du, J., Estrella, M., Solorio-Kirpichyan, K., Jeffrey, P.D., Korennykh, A.(2022) Proc Natl Acad Sci U S A 119: e2200923119-e2200923119
- PubMed: 35733246
- DOI: https://doi.org/10.1073/pnas.2200923119
- Primary Citation of Related Structures:
7N29 - PubMed Abstract:
All kingdoms of life produce essential nicotinamide dinucleotide NADP(H) using NAD kinases (NADKs). A panel of published NADK structures from bacteria, eukaryotic cytosol, and yeast mitochondria revealed similar tetrameric enzymes. Here, we present the 2.8-Å structure of the human mitochondrial kinase NADK2 with a bound substrate, which is an exception to this uniformity, diverging both structurally and biochemically from NADKs. We show that NADK2 harbors a unique tetramer disruptor/dimerization e lement, which is conserved in m itochondrial k inases of a nimals (EMKA) and absent from other NADKs. EMKA stabilizes the NADK2 dimer but prevents further NADK2 oligomerization by blocking the tetramerization interface. This structural change bears functional consequences and alters the activation mechanism of the enzyme. Whereas tetrameric NADKs undergo cooperative activation via oligomerization, NADK2 is a constitutively active noncooperative dimer. Thus, our data point to a unique regulation of NADP(H) synthesis in animal mitochondria achieved via structural adaptation of the NADK2 kinase.
Organizational Affiliation:
Department of Molecular Biology, Princeton University, Princeton, NJ 08544.