Structural assessment of HLA-A2-restricted SARS-CoV-2 spike epitopes recognized by public and private T-cell receptors.
Wu, D., Kolesnikov, A., Yin, R., Guest, J.D., Gowthaman, R., Shmelev, A., Serdyuk, Y., Dianov, D.V., Efimov, G.A., Pierce, B.G., Mariuzza, R.A.(2022) Nat Commun 13: 19-19
- PubMed: 35013235
- DOI: https://doi.org/10.1038/s41467-021-27669-8
- Primary Citation of Related Structures:
7N1A, 7N1B, 7N1C, 7N1D, 7N1E, 7N1F - PubMed Abstract:
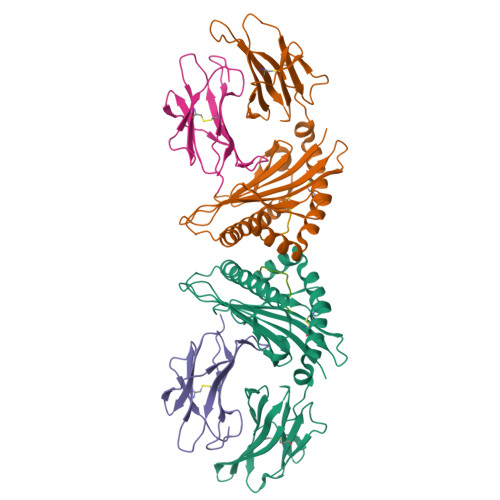
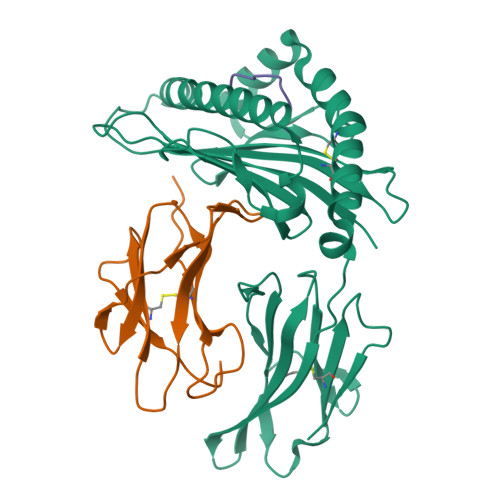
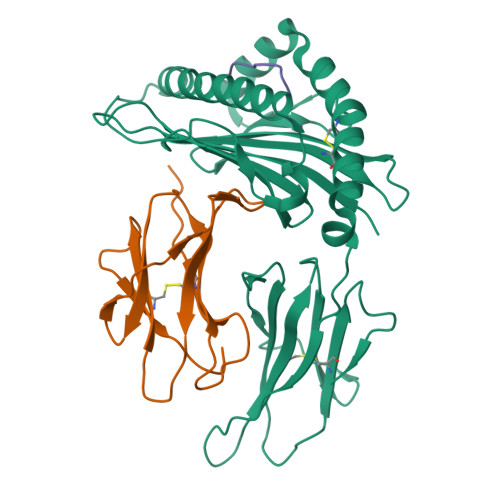
T cells play a vital role in combatting SARS-CoV-2 and forming long-term memory responses. Whereas extensive structural information is available on neutralizing antibodies against SARS-CoV-2, such information on SARS-CoV-2-specific T-cell receptors (TCRs) bound to their peptide-MHC targets is lacking. Here we determine the structures of a public and a private TCR from COVID-19 convalescent patients in complex with HLA-A2 and two SARS-CoV-2 spike protein epitopes (YLQ and RLQ). The structures reveal the basis for selection of particular TRAV and TRBV germline genes by the public but not the private TCR, and for the ability of the TCRs to recognize natural variants of RLQ but not YLQ. Neither TCR recognizes homologous epitopes from human seasonal coronaviruses. By elucidating the mechanism for TCR recognition of an immunodominant yet variable epitope (YLQ) and a conserved but less commonly targeted epitope (RLQ), this study can inform prospective efforts to design vaccines to elicit pan-coronavirus immunity.
Organizational Affiliation:
W.M. Keck Laboratory for Structural Biology, University of Maryland Institute for Bioscience and Biotechnology Research, Rockville, MD, 20850, USA.