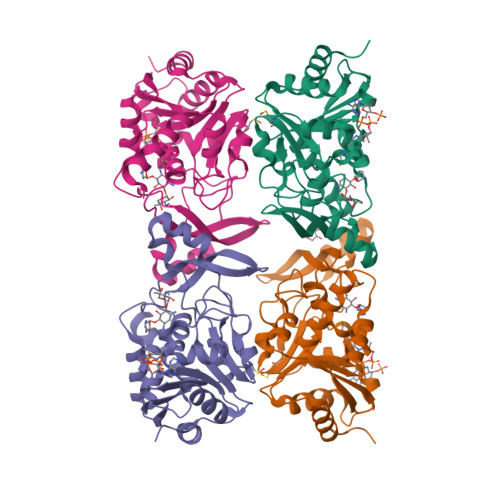
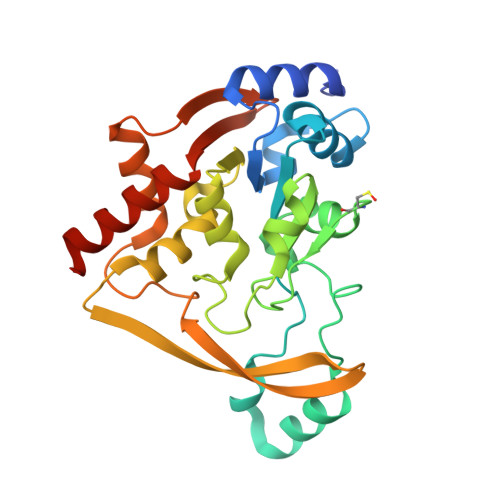
Structural elucidation of substrate-bound aminoglycoside acetyltransferase (3)-IIIa.
Zielinski, M., Blanchet, J., Hailemariam, S., Berghuis, A.M.(2022) PLoS One 17: e0269684-e0269684
- PubMed: 35921328
- DOI: https://doi.org/10.1371/journal.pone.0269684
- Primary Citation of Related Structures:
7MQK, 7MQL, 7MQM - PubMed Abstract:
Canonical aminoglycosides are a large group of antibiotics, where the part of chemical diversity stems from the substitution of the neamine ring system on positions 5 and 6. Certain aminoglycoside modifying enzymes can modify a broad range of 4,5- and 4,6-disubstituted aminoglycosides, with some as many as 15. This study presents the structural and kinetic results describing a promiscuous aminoglycoside acetyltransferase AAC(3)-IIIa. This enzyme has been crystallized in ternary complex with coenzyme A and 4,5- and 4,6-disubstituted aminoglycosides. We have followed up this work with kinetic characterization utilizing a panel of diverse aminoglycosides, including a next-generation aminoglycoside, plazomicin. Lastly, we observed an alternative binding mode of gentamicin in the aminoglycoside binding site, which was proven to be a crystallographic artifact based on mutagenesis.
Organizational Affiliation:
Department of Biochemistry, McGill University, Montréal, Québec, Canada.