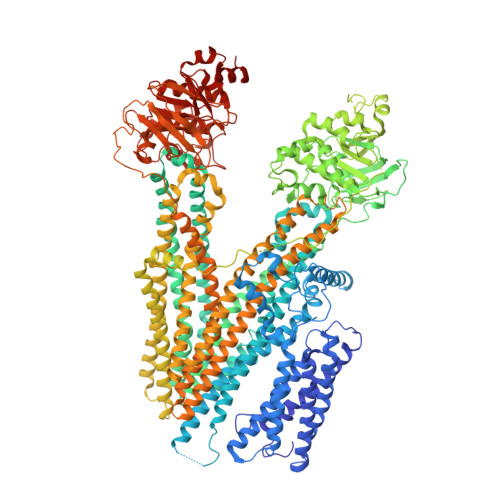
Structure of Ycf1p reveals the transmembrane domain TMD0 and the regulatory region of ABCC transporters.
Bickers, S.C., Benlekbir, S., Rubinstein, J.L., Kanelis, V.(2021) Proc Natl Acad Sci U S A 118
- PubMed: 34021087
- DOI: https://doi.org/10.1073/pnas.2025853118
- Primary Citation of Related Structures:
7MPE - PubMed Abstract:
ATP binding cassette (ABC) proteins typically function in active transport of solutes across membranes. The ABC core structure is composed of two transmembrane domains (TMD1 and TMD2) and two cytosolic nucleotide binding domains (NBD1 and NBD2). Some members of the C-subfamily of ABC (ABCC) proteins, including human multidrug resistance proteins (MRPs), also possess an N-terminal transmembrane domain (TMD0) that contains five transmembrane α-helices and is connected to the ABC core by the L0 linker. While TMD0 was resolved in SUR1, the atypical ABCC protein that is part of the hetero-octameric ATP-sensitive K + channel, little is known about the structure of TMD0 in monomeric ABC transporters. Here, we present the structure of yeast cadmium factor 1 protein (Ycf1p), a homolog of human MRP1, determined by electron cryo-microscopy (cryo-EM). A comparison of Ycf1p, SUR1, and a structure of MRP1 that showed TMD0 at low resolution demonstrates that TMD0 can adopt different orientations relative to the ABC core, including a ∼145° rotation between Ycf1p and SUR1. The cryo-EM map also reveals that segments of the regulatory (R) region, which links NBD1 to TMD2 and was poorly resolved in earlier ABCC structures, interacts with the L0 linker, NBD1, and TMD2. These interactions, combined with fluorescence quenching experiments of isolated NBD1 with and without the R region, suggest how posttranslational modifications of the R region modulate ABC protein activity. Mapping known mutations from MRP2 and MRP6 onto the Ycf1p structure explains how mutations involving TMD0 and the R region of these proteins lead to disease.
Organizational Affiliation:
Department of Chemistry, University of Toronto, Toronto, ON M5S 3H6, Canada.