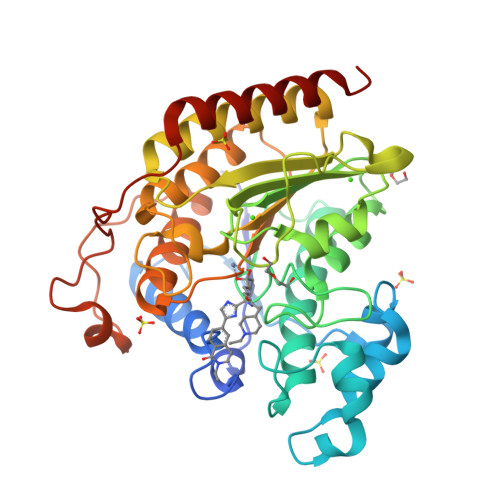
Discovery of macrocyclic HDACs 1, 2, and 3 selective inhibitors for HIV latency reactivation.
Yu, W., Fells, J., Clausen, D., Liu, J., Klein, D.J., Christine Chung, C., Myers, R.W., Wu, J., Wu, G., Howell, B.J., Barnard, R.J.O., Kozlowski, J.(2021) Bioorg Med Chem Lett 47: 128168-128168
- PubMed: 34091041
- DOI: https://doi.org/10.1016/j.bmcl.2021.128168
- Primary Citation of Related Structures:
7MOS, 7MOT, 7MOX, 7MOY, 7MOZ - PubMed Abstract:
A series of unique macrocyclic HDACs 1, 2, and 3 selective inhibitors were identified with good enzymatic activity and high selectivity over HDACs 6 and 8. These macrocyclic HDAC inhibitors used an ethyl ketone as the zinc-binding group. Compounds 25 and 26 stood out as leads due to their low double-digit nM EC 50 s in the 2C4 cell-based HIV latency reactivation assay. The PK profiles of these macrocyclic HDAC inhibitors still needed improvement.
Organizational Affiliation:
Merck & Co., Inc., 2000 Galloping Hill Road, Kenilworth, NJ 07033, USA. Electronic address: wenshengyu@msn.com.