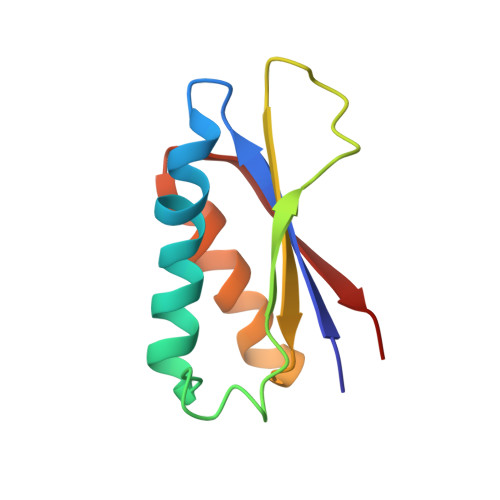
Design and characterization of a protein fold switching network.
Ruan, B., He, Y., Chen, Y., Choi, E.J., Chen, Y., Motabar, D., Solomon, T., Simmerman, R., Kauffman, T., Gallagher, D.T., Orban, J., Bryan, P.N.(2023) Nat Commun 14: 431-431
- PubMed: 36702827
- DOI: https://doi.org/10.1038/s41467-023-36065-3
- Primary Citation of Related Structures:
7MN1, 7MN2, 7MP7, 7MQ4 - PubMed Abstract:
To better understand how amino acid sequence encodes protein structure, we engineered mutational pathways that connect three common folds (3α, β-grasp, and α/β-plait). The structures of proteins at high sequence-identity intersections in the pathways (nodes) were determined using NMR spectroscopy and analyzed for stability and function. To generate nodes, the amino acid sequence encoding a smaller fold is embedded in the structure of an ~50% larger fold and a new sequence compatible with two sets of native interactions is designed. This generates protein pairs with a 3α or β-grasp fold in the smaller form but an α/β-plait fold in the larger form. Further, embedding smaller antagonistic folds creates critical states in the larger folds such that single amino acid substitutions can switch both their fold and function. The results help explain the underlying ambiguity in the protein folding code and show that new protein structures can evolve via abrupt fold switching.
Organizational Affiliation:
Potomac Affinity Proteins, 11305 Dunleith Pl, North Potomac, MD, 20878, USA.