Cross-HLA targeting of intracellular oncoproteins with peptide-centric CARs.
Yarmarkovich, M., Marshall, Q.F., Warrington, J.M., Premaratne, R., Farrel, A., Groff, D., Li, W., di Marco, M., Runbeck, E., Truong, H., Toor, J.S., Tripathi, S., Nguyen, S., Shen, H., Noel, T., Church, N.L., Weiner, A., Kendsersky, N., Martinez, D., Weisberg, R., Christie, M., Eisenlohr, L., Bosse, K.R., Dimitrov, D.S., Stevanovic, S., Sgourakis, N.G., Kiefel, B.R., Maris, J.M.(2021) Nature 599: 477-484
- PubMed: 34732890
- DOI: https://doi.org/10.1038/s41586-021-04061-6
- Primary Citation of Related Structures:
7MJ6, 7MJ7, 7MJ8, 7MJ9, 7MJA - PubMed Abstract:
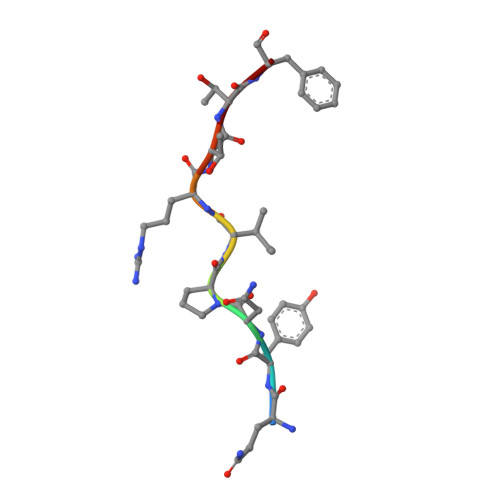
The majority of oncogenic drivers are intracellular proteins, thus constraining their immunotherapeutic targeting to mutated peptides (neoantigens) presented by individual human leukocyte antigen (HLA) allotypes 1 . However, most cancers have a modest mutational burden that is insufficient to generate responses using neoantigen-based therapies 2,3 . Neuroblastoma is a paediatric cancer that harbours few mutations and is instead driven by epigenetically deregulated transcriptional networks 4 . Here we show that the neuroblastoma immunopeptidome is enriched with peptides derived from proteins that are essential for tumourigenesis and focus on targeting the unmutated peptide QYNPIRTTF, discovered on HLA-A*24:02, which is derived from the neuroblastoma dependency gene and master transcriptional regulator PHOX2B. To target QYNPIRTTF, we developed peptide-centric chimeric antigen receptors (CARs) using a counter-panning strategy with predicted potentially cross-reactive peptides. We further hypothesized that peptide-centric CARs could recognize peptides on additional HLA allotypes when presented in a similar manner. Informed by computational modelling, we showed that PHOX2B peptide-centric CARs also recognize QYNPIRTTF presented by HLA-A*23:01 and the highly divergent HLA-B*14:02. Finally, we demonstrated potent and specific killing of neuroblastoma cells expressing these HLAs in vitro and complete tumour regression in mice. These data suggest that peptide-centric CARs have the potential to vastly expand the pool of immunotherapeutic targets to include non-immunogenic intracellular oncoproteins and widen the population of patients who would benefit from such therapy by breaking conventional HLA restriction.
Organizational Affiliation:
Division of Oncology and Center for Childhood Cancer Research, Children's Hospital of Philadelphia, Philadelphia, PA, USA.