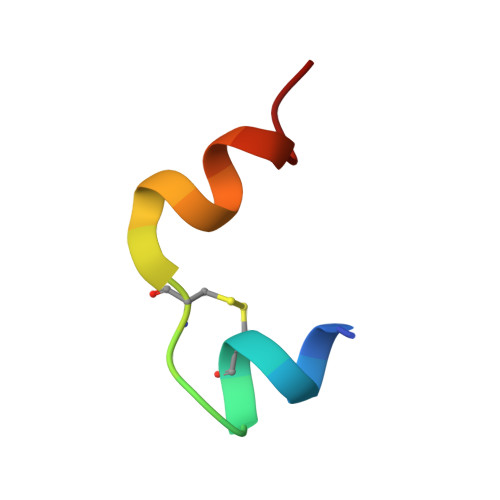Functionally selective signaling and broad metabolic benefits by novel insulin receptor partial agonists.
Wu, M., Carballo-Jane, E., Zhou, H., Zafian, P., Dai, G., Liu, M., Lao, J., Kelly, T., Shao, D., Gorski, J., Pissarnitski, D., Kekec, A., Chen, Y., Previs, S.F., Scapin, G., Llorente, Y.G., Hollingsworth, S.A., Yan, L., Feng, D., Huo, P., Walford, G., Erion, M.D., Kelley, D.E., Lin, S., Mu, J.(2022) Nat Commun 13: 942-942
- PubMed: 35177603
- DOI: https://doi.org/10.1038/s41467-022-28561-9
- Primary Citation of Related Structures:
7MD4, 7MD5 - PubMed Abstract:
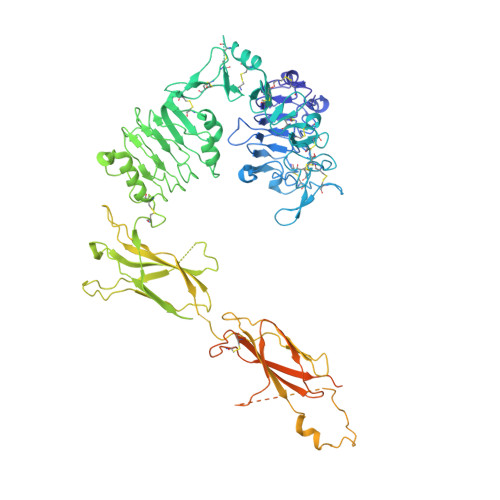
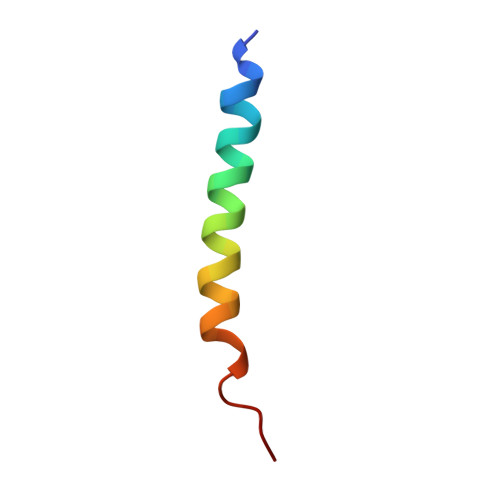
Insulin analogs have been developed to treat diabetes with focus primarily on improving the time action profile without affecting ligand-receptor interaction or functional selectivity. As a result, inherent liabilities (e.g. hypoglycemia) of injectable insulin continue to limit the true therapeutic potential of related agents. Insulin dimers were synthesized to investigate whether partial agonism of the insulin receptor (IR) tyrosine kinase is achievable, and to explore the potential for tissue-selective systemic insulin pharmacology. The insulin dimers induced distinct IR conformational changes compared to native monomeric insulin and substrate phosphorylation assays demonstrated partial agonism. Structurally distinct dimers with differences in conjugation sites and linkers were prepared to deliver desirable IR partial agonist (IRPA). Systemic infusions of a B29-B29 dimer in vivo revealed sharp differences compared to native insulin. Suppression of hepatic glucose production and lipolysis were like that attained with regular insulin, albeit with a distinctly shallower dose-response. In contrast, there was highly attenuated stimulation of glucose uptake into muscle. Mechanistic studies indicated that IRPAs exploit tissue differences in receptor density and have additional distinctions pertaining to drug clearance and distribution. The hepato-adipose selective action of IRPAs is a potentially safer approach for treatment of diabetes.
Organizational Affiliation:
Merck & Co., Inc., Kenilworth, NJ, 07033, USA.