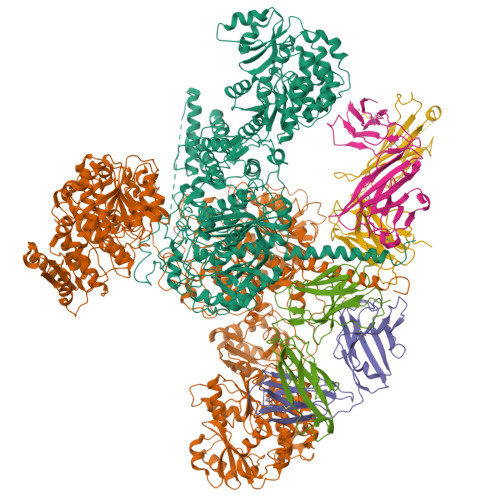
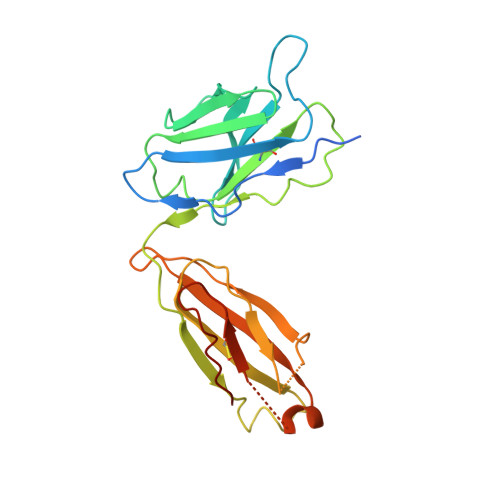
Mapping the catalytic conformations of an assembly-line polyketide synthase module.
Cogan, D.P., Zhang, K., Li, X., Li, S., Pintilie, G.D., Roh, S.H., Craik, C.S., Chiu, W., Khosla, C.(2021) Science 374: 729-734
- PubMed: 34735239
- DOI: https://doi.org/10.1126/science.abi8358
- Primary Citation of Related Structures:
7M7E, 7M7F, 7M7G, 7M7H, 7M7I, 7M7J - PubMed Abstract:
Assembly-line polyketide synthases, such as the 6-deoxyerythronolide B synthase (DEBS), are large enzyme factories prized for their ability to produce specific and complex polyketide products. By channeling protein-tethered substrates across multiple active sites in a defined linear sequence, these enzymes facilitate programmed small-molecule syntheses that could theoretically be harnessed to access countless polyketide product structures. Using cryogenic electron microscopy to study DEBS module 1, we present a structural model describing this substrate-channeling phenomenon. Our 3.2- to 4.3-angstrom-resolution structures of the intact module reveal key domain-domain interfaces and highlight an unexpected module asymmetry. We also present the structure of a product-bound module that shines light on a recently described “turnstile” mechanism for transient gating of active sites along the assembly line.
Organizational Affiliation:
Department of Chemistry, Stanford University, Stanford, CA 94305, USA.