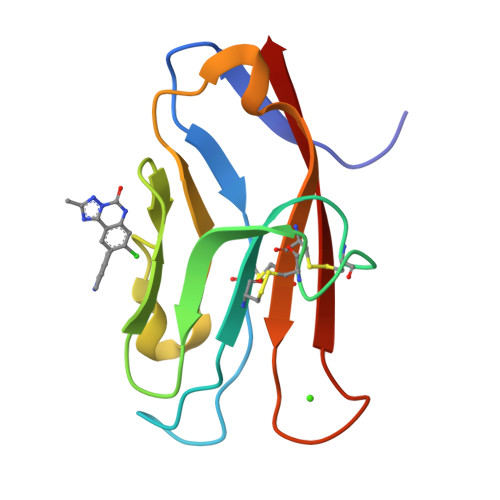
Fragment-Based Discovery of Small Molecules Bound to T-Cell Immunoglobulin and Mucin Domain-Containing Molecule 3 (TIM-3).
Rietz, T.A., Teuscher, K.B., Mills, J.J., Gogliotti, R.D., Lepovitz, L.T., Scaggs, W.R., Yoshida, K., Luong, K., Lee, T., Fesik, S.W.(2021) J Med Chem 64: 14757-14772
- PubMed: 34597046
- DOI: https://doi.org/10.1021/acs.jmedchem.1c01336
- Primary Citation of Related Structures:
7M3Y, 7M3Z, 7M41 - PubMed Abstract:
T-cell immunoglobulin and mucin domain-containing molecule 3 (TIM-3; HAVCR2) has emerged as an attractive immune checkpoint target for cancer immunotherapy. TIM-3 is a negative regulator of the systemic immune response to cancer and is expressed on several dysfunctional, or exhausted, immune cell subsets. Upregulation of TIM-3 is associated with tumor progression, poor survival rates, and acquired resistance to antibody-based immunotherapies in the clinic. Despite the potential advantages of small-molecule inhibitors over antibodies, the discovery of small-molecule inhibitors has lagged behind that of antibody therapeutics. Here, we describe the discovery of high-affinity small-molecule ligands for TIM-3 through an NMR-based fragment screen and structure-based lead optimization. These compounds represent useful tools to further study the biology of TIM-3 immune modulation in cancer and serve as a potentially useful starting point toward the discovery of TIM-3-targeted therapeutics.
Organizational Affiliation:
Department of Biochemistry, Vanderbilt University School of Medicine, Nashville, Tennessee 37232-0146, United States.