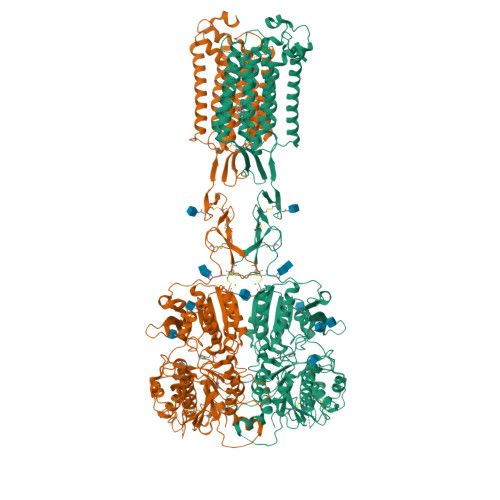
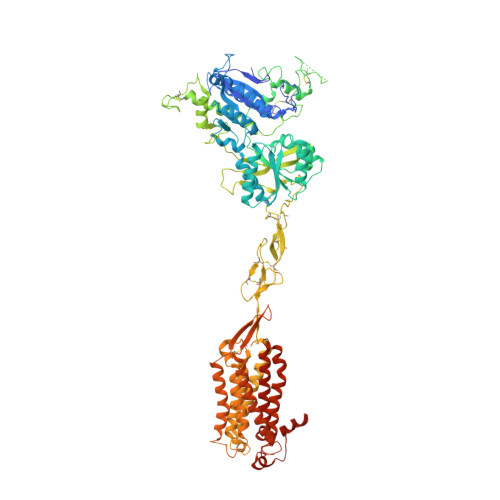
Asymmetric activation of the calcium-sensing receptor homodimer.
Gao, Y., Robertson, M.J., Rahman, S.N., Seven, A.B., Zhang, C., Meyerowitz, J.G., Panova, O., Hannan, F.M., Thakker, R.V., Brauner-Osborne, H., Mathiesen, J.M., Skiniotis, G.(2021) Nature 595: 455-459
- PubMed: 34194040
- DOI: https://doi.org/10.1038/s41586-021-03691-0
- Primary Citation of Related Structures:
7M3E, 7M3F, 7M3G, 7M3J - PubMed Abstract:
The calcium-sensing receptor (CaSR), a cell-surface sensor for Ca 2+ , is the master regulator of calcium homeostasis in humans and is the target of calcimimetic drugs for the treatment of parathyroid disorders 1 . CaSR is a family C G-protein-coupled receptor 2 that functions as an obligate homodimer, with each protomer composed of a Ca 2+ -binding extracellular domain and a seven-transmembrane-helix domain (7TM) that activates heterotrimeric G proteins. Here we present cryo-electron microscopy structures of near-full-length human CaSR in inactive or active states bound to Ca 2+ and various calcilytic or calcimimetic drug molecules. We show that, upon activation, the CaSR homodimer adopts an asymmetric 7TM configuration that primes one protomer for G-protein coupling. This asymmetry is stabilized by 7TM-targeting calcimimetic drugs adopting distinctly different poses in the two protomers, whereas the binding of a calcilytic drug locks CaSR 7TMs in an inactive symmetric configuration. These results provide a detailed structural framework for CaSR activation and the rational design of therapeutics targeting this receptor.
Organizational Affiliation:
Department of Molecular and Cellular Physiology, Stanford University School of Medicine, Stanford, CA, USA.