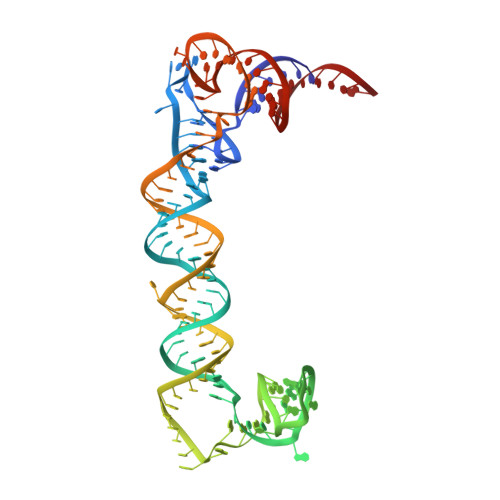
Structures of flavivirus RNA promoters suggest two binding modes with NS5 polymerase.
Lee, E., Bujalowski, P.J., Teramoto, T., Gottipati, K., Scott, S.D., Padmanabhan, R., Choi, K.H.(2021) Nat Commun 12: 2530-2530
- PubMed: 33953197
- DOI: https://doi.org/10.1038/s41467-021-22846-1
- Primary Citation of Related Structures:
7LYF, 7LYG - PubMed Abstract:
Flaviviruses use a ~70 nucleotide stem-loop structure called stem-loop A (SLA) at the 5' end of the RNA genome as a promoter for RNA synthesis. Flaviviral polymerase NS5 specifically recognizes SLA to initiate RNA synthesis and methylate the 5' guanosine cap. We report the crystal structures of dengue (DENV) and Zika virus (ZIKV) SLAs. DENV and ZIKV SLAs differ in the relative orientations of their top stem-loop helices to bottom stems, but both form an intermolecular three-way junction with a neighboring SLA molecule. To understand how NS5 engages SLA, we determined the SLA-binding site on NS5 and modeled the NS5-SLA complex of DENV and ZIKV. Our results show that the gross conformational differences seen in DENV and ZIKV SLAs can be compensated by the differences in the domain arrangements in DENV and ZIKV NS5s. We describe two binding modes of SLA and NS5 and propose an SLA-mediated RNA synthesis mechanism.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, Sealy Center for Structural Biology and Molecular Biophysics, The University of Texas Medical Branch, 301 University Boulevard, Galveston, TX, USA.