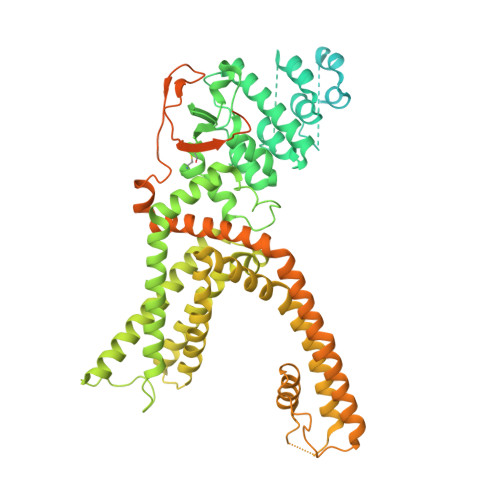
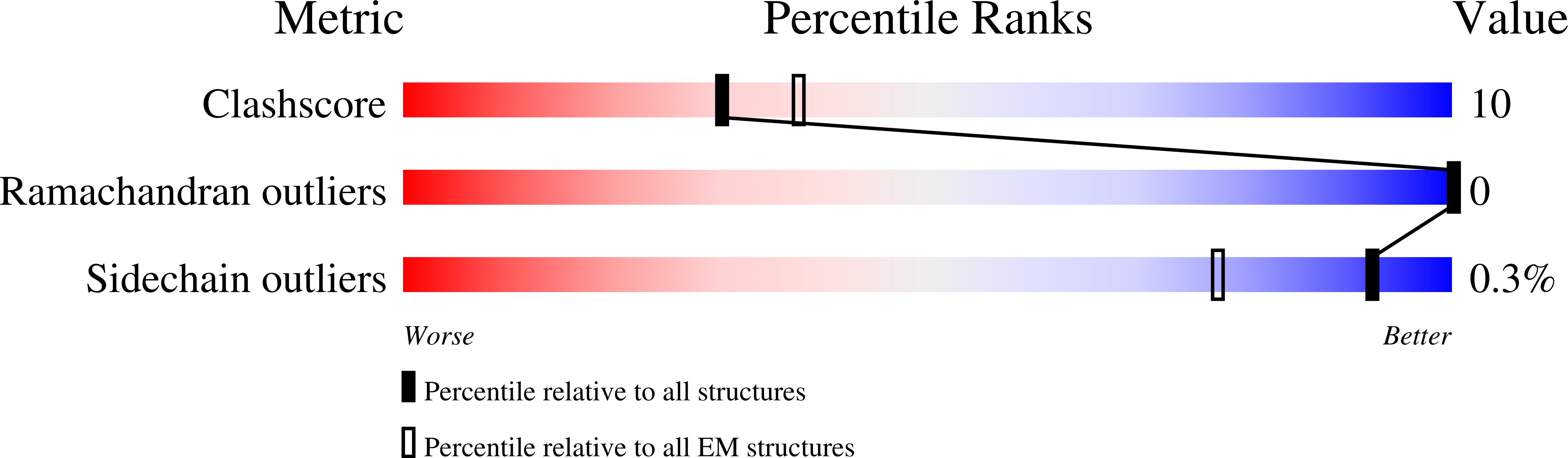Heat-dependent opening of TRPV1 in the presence of capsaicin.
Kwon, D.H., Zhang, F., Suo, Y., Bouvette, J., Borgnia, M.J., Lee, S.Y.(2021) Nat Struct Mol Biol 28: 554-563
- PubMed: 34239123
- DOI: https://doi.org/10.1038/s41594-021-00616-3
- Primary Citation of Related Structures:
7LP9, 7LPA, 7LPB, 7LPC, 7LPD, 7LPE - PubMed Abstract:
Transient receptor potential vanilloid member 1 (TRPV1) is a Ca 2+ -permeable cation channel that serves as the primary heat and capsaicin sensor in humans. Using cryo-EM, we have determined the structures of apo and capsaicin-bound full-length rat TRPV1 reconstituted into lipid nanodiscs over a range of temperatures. This has allowed us to visualize the noxious heat-induced opening of TRPV1 in the presence of capsaicin. Notably, noxious heat-dependent TRPV1 opening comprises stepwise conformational transitions. Global conformational changes across multiple subdomains of TRPV1 are followed by the rearrangement of the outer pore, leading to gate opening. Solvent-accessible surface area analyses and functional studies suggest that a subset of residues form an interaction network that is directly involved in heat sensing. Our study provides a glimpse of the molecular principles underlying noxious physical and chemical stimuli sensing by TRPV1, which can be extended to other thermal sensing ion channels.
Organizational Affiliation:
Department of Biochemistry, Duke University School of Medicine, Durham, NC, USA.