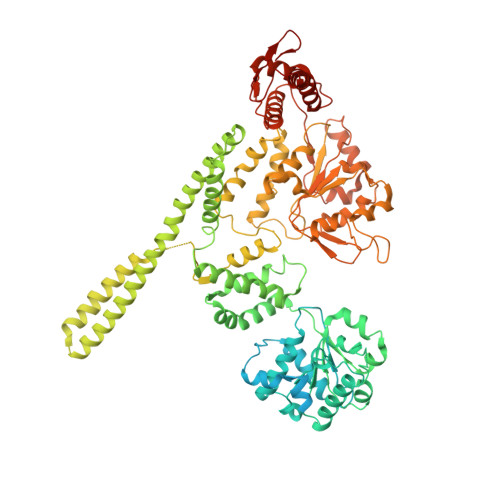
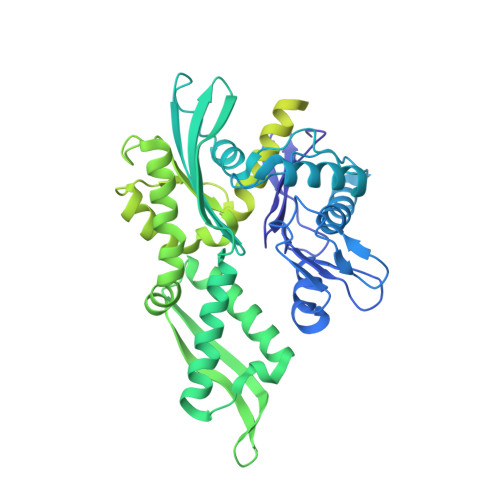
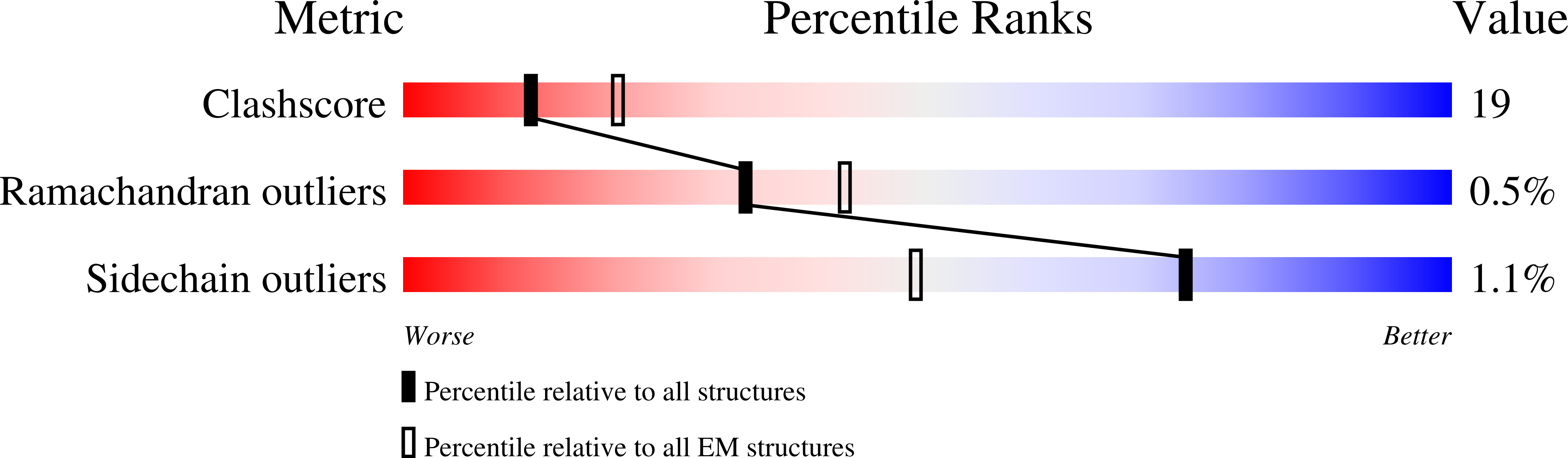Structural basis for aggregate dissolution and refolding by the Mycobacterium tuberculosis ClpB-DnaK bi-chaperone system.
Yin, Y., Feng, X., Yu, H., Fay, A., Kovach, A., Glickman, M.S., Li, H.(2021) Cell Rep 35: 109166-109166
- PubMed: 34038719
- DOI: https://doi.org/10.1016/j.celrep.2021.109166
- Primary Citation of Related Structures:
6W6E, 6W6G, 6W6H, 6W6I, 6W6J, 7L6N - PubMed Abstract:
The M. tuberculosis (Mtb) ClpB is a protein disaggregase that helps to rejuvenate the bacterial cell. DnaK is a protein foldase that can function alone, but it can also bind to the ClpB hexamer to physically couple protein disaggregation with protein refolding, although the molecular mechanism is not well understood. Here, we report the cryo-EM analysis of the Mtb ClpB-DnaK bi-chaperone in the presence of ATPγS and a protein substrate. We observe three ClpB conformations in the presence of DnaK, identify a conserved TGIP loop linking the oligonucleotide/oligosaccharide-binding domain and the nucleotide-binding domain that is important for ClpB function, derive the interface between the regulatory middle domain of the ClpB and the DnaK nucleotide-binding domain, and find that DnaK binding stabilizes, but does not bend or tilt, the ClpB middle domain. We propose a model for the synergistic actions of aggregate dissolution and refolding by the Mtb ClpB-DnaK bi-chaperone system.
Organizational Affiliation:
Department of Structural Biology, Van Andel Institute, Grand Rapids, MI, USA.