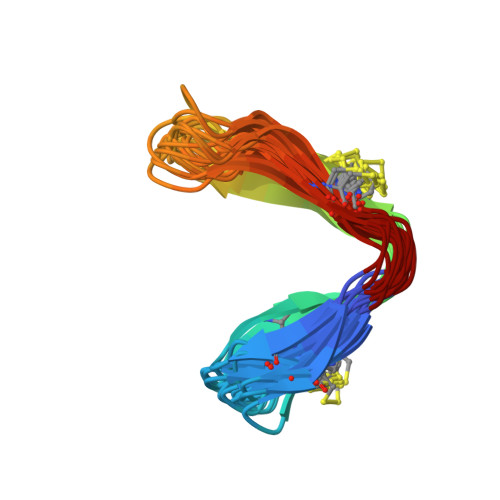
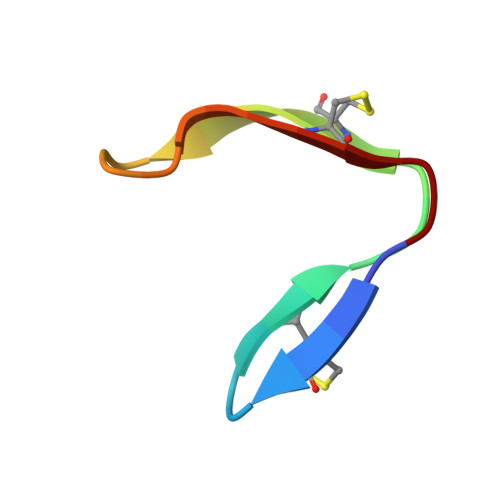
A chameleonic macrocyclic peptide with drug delivery applications.
Payne, C.D., Franke, B., Fisher, M.F., Hajiaghaalipour, F., McAleese, C.E., Song, A., Eliasson, C., Zhang, J., Jayasena, A.S., Vadlamani, G., Clark, R.J., Minchin, R.F., Mylne, J.S., Rosengren, K.J.(2021) Chem Sci 12: 6670-6683
- PubMed: 34040741
- DOI: https://doi.org/10.1039/d1sc00692d
- Primary Citation of Related Structures:
7L51, 7L53, 7L54, 7L55 - PubMed Abstract:
Head-to-tail cyclized peptides are intriguing natural products with unusual properties. The PawS-Derived Peptides (PDPs) are ribosomally synthesized as part of precursors for seed storage albumins in species of the daisy family, and are post-translationally excised and cyclized during proteolytic processing. Here we report a PDP twice the typical size and with two disulfide bonds, identified from seeds of Zinnia elegans . In water, synthetic PDP-23 forms a unique dimeric structure in which two monomers containing two β-hairpins cross-clasp and enclose a hydrophobic core, creating a square prism. This dimer can be split by addition of micelles or organic solvent and in monomeric form PDP-23 adopts open or closed V-shapes, exposing different levels of hydrophobicity dependent on conditions. This chameleonic character is unusual for disulfide-rich peptides and engenders PDP-23 with potential for cell delivery and accessing novel targets. We demonstrate this by conjugating a rhodamine dye to PDP-23, creating a stable, cell-penetrating inhibitor of the P-glycoprotein drug efflux pump.
Organizational Affiliation:
The University of Queensland, School of Biomedical Sciences Brisbane QLD 4072 Australia j.rosengren@uq.edu.au.