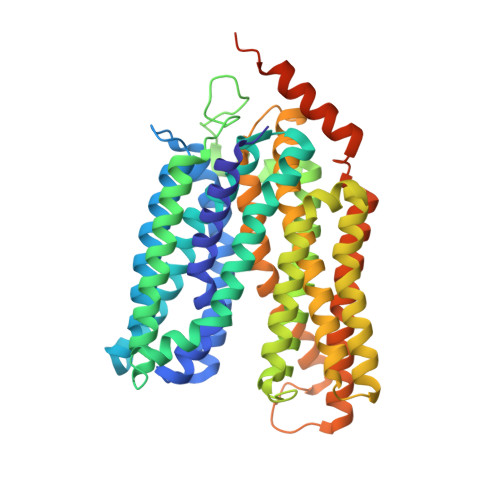
X-ray crystallography reveals molecular recognition mechanism for sugar binding in a melibiose transporter MelB.
Guan, L., Hariharan, P.(2021) Commun Biol 4: 931-931
- PubMed: 34341464
- DOI: https://doi.org/10.1038/s42003-021-02462-x
- Primary Citation of Related Structures:
7L16, 7L17 - PubMed Abstract:
Major facilitator superfamily_2 transporters are widely found from bacteria to mammals. The melibiose transporter MelB, which catalyzes melibiose symport with either Na + , Li + , or H + , is a prototype of the Na + -coupled MFS transporters, but its sugar recognition mechanism has been a long-unsolved puzzle. Two high-resolution X-ray crystal structures of a Salmonella typhimurium MelB mutant with a bound ligand, either nitrophenyl-α-D-galactoside or dodecyl-β-D-melibioside, were refined to a resolution of 3.05 or 3.15 Å, respectively. In the substrate-binding site, the interaction of both galactosyl moieties on the two ligands with MelB St are virturally same, so the sugar specificity determinant pocket can be recognized, and hence the molecular recognition mechanism for sugar binding in MelB has been deciphered. The conserved cation-binding pocket is also proposed, which directly connects to the sugar specificity pocket. These key structural findings have laid a solid foundation for our understanding of the cooperative binding and symport mechanisms in Na + -coupled MFS transporters, including eukaryotic transporters such as MFSD2A.
Organizational Affiliation:
Department of Cell Physiology and Molecular Biophysics, Center for Membrane Protein Research, School of Medicine, Texas Tech University Health Sciences Center, Lubbock, TX, USA. Lan.Guan@ttuhsc.edu.